The global healthcare system and economy have suffered massively throughout the ongoing coronavirus disease 2019 (COVID-19) pandemic, which emerged as a result of the rapid spread of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Owing to the evolution of the virus due to mutations, several SARS-CoV-2 variants have emerged, some of which are more virulent and transmissible than the original strain.
SARS-CoV-2 variants have been classified as variants of concern (VOC) and variants of interest (VOI) by the World Health Organization (WHO) based on their increased virulence, changes in epidemiology and/or clinical presentation, increased transmissibility and decreased effectiveness of social measures or COVID-19 diagnostics. Recently, the SARS-CoV-2 Omicron variant has become the dominantly circulating strain that replaced the previously dominant Delta variant in many countries across the world.
.jpg)
Study: Omicron BA.1 and BA.2 are antigenically distinct SARS-CoV-2 variants. Image Credit: CROCOTHERY / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
To date, all available COVID-19 vaccines and therapeutics have been developed against the spike (S) protein of the original SARS-CoV-2 strain. Therefore, mutations in the S domain could pose a threat to the effectiveness of current COVID-19 vaccines.
Previous studies have reported that the efficacy of existing COVID-19 vaccines is reduced against some SARS-CoV-2 variants, such as the Omicron variant. Furthermore, several reports have also found that the Omicron BA.1 variant almost completely escapes neutralizing antibodies, which has resulted in a high number of breakthrough infections. However, the BA.1 lineage has been determined to be less virulent as compared to other SARS-CoV-2 variants.
According to recent studies, Omicron variants BA.2 and BA.1 are genetically different. In many countries, BA.1 has already been replaced by the new BA.2 strain. To date, no study on the antigenic characteristics of BA.2 and BA.1 is available.
A new study published on the bioRxiv* preprint server identifies the antigenic differences that exist between the Omicron BA.2 and BA.1 variants. Herein, the researchers also quantified the extent to which human post-BNT162b2 immunization sera neutralized Omicron BA.2.
Study findings
In this study, researchers used antigenic cartography to estimate and visualize antigenic differences between SARS-CoV-2 variants using hamster sera, which was collected after primary infection. Scientists found it difficult to obtain human sera post-primary Omicron infection; therefore, they selected Syrian golden hamsters’ sera for this study.
Hamsters are highly susceptible to SARS-CoV-2 infection and are ideal for collecting well-defined sera. Furthermore, previous studies have reported that human and hamster serum responses to SARS-CoV-2 infection are similar, as they develop similar topological maps.
In the current study, scientists inoculated hamsters with several SARS-CoV-2 variants including 614G, Alpha, Beta, Gamma, Zeta, Delta, Mu, and Omicron (BA.1) to generate antisera.
Antigenic cartography allows for the quantitative analysis of antigenic drift and visualization of new antigenic clusters. Utilizing this technique, early SARS-CoV-2 variants have been found to be antigenically similar, as they were found to cluster closer to each other in antigenic space. This is comparable to Omicron BA.1 and BA.2, which have evolved as two distinct antigenic outliers.
Plaque reduction neutralization titers resulted in a 50% reduction in infected cells associated with authentic SARS-CoV-2, as well as pseudotyped viral infection. The researchers reported that sera from Omicron BA.1-infected hamsters poorly neutralized all other variants, which implies that this variant induces different antibody responses as compared to other SARS-CoV-2 variants.
Antigenic maps were developed by a multidimensional scaling algorithm. To this end, the scientists reported that the map constructed by Calu-3 cells was highly similar to the VeroE6 map because the same antigens were plotted within one two-fold dilution from each other in the two maps. This finding demonstrates that the choice of the cell line for the neutralization assay had no substantial effect on the topology of the map.
The Omicron BA.1 formed a distinct antigenic outlier in the map that was 10- to 38-fold dilutions away from the nearest virus.
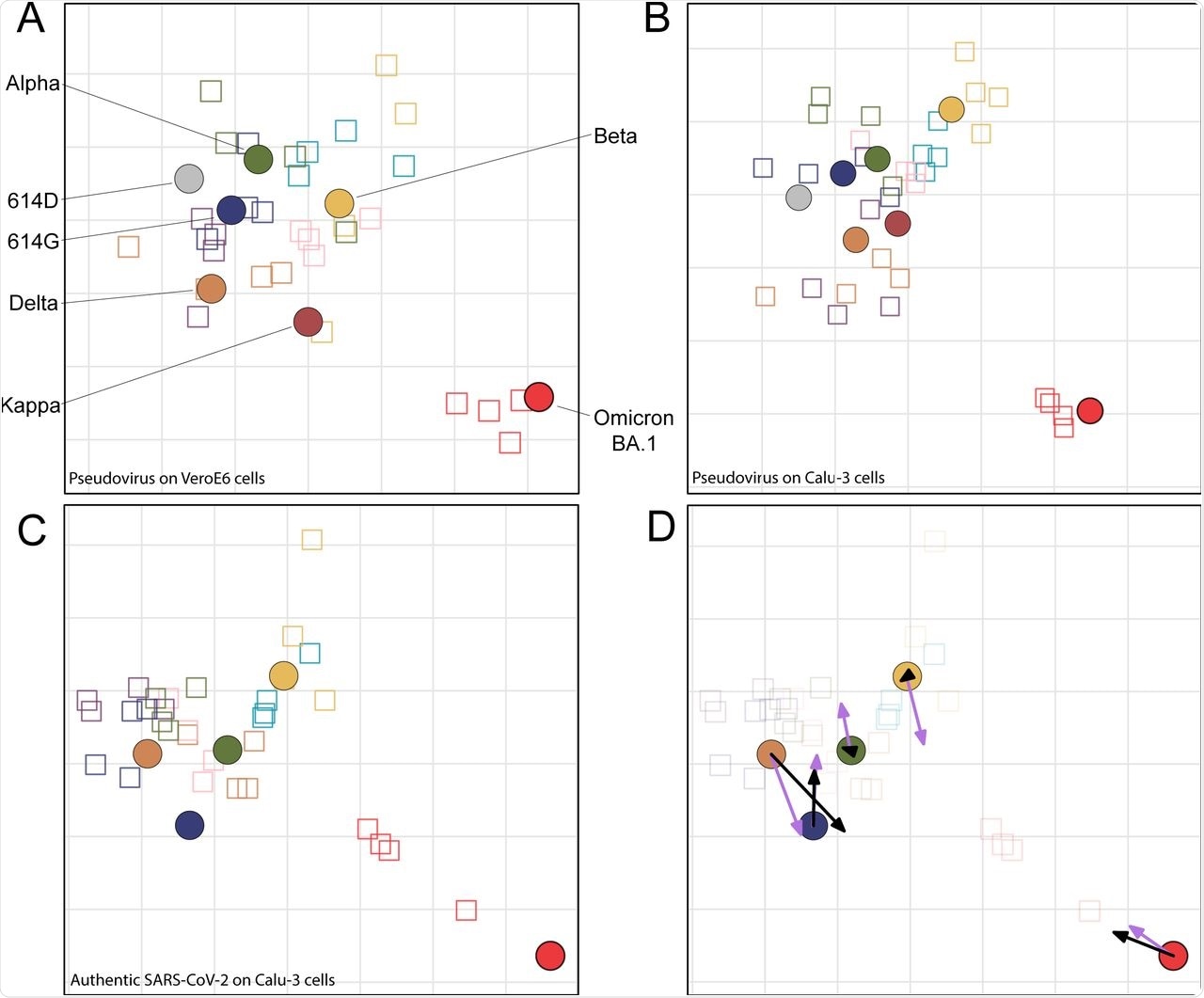 Antigenic maps Comparing neutralizations with SARS-CoV-2 pseudoviruses and authentic SARS-CoV-2.
Antigenic maps Comparing neutralizations with SARS-CoV-2 pseudoviruses and authentic SARS-CoV-2.
The researchers also reported that a single BNT162b2 vaccination exhibited a low neutralizing titer against all variants with an addition 13-fold and 8-fold reduction in neutralizing titers observed against Omicron BA.1 and BA.2, respectively. Both Omicron variants were also found to evade vaccine-induced antibody responses as a result of their unique antigenic characteristics.
Conclusions
The emergence and rapid spread of the highly mutated BA.1 and BA.2 variants was an indicator that population immunity is employing robust selective pressure on SARS-CoV-2 in favor of the emergence of new antigenic variants.
The researchers of the current study anticipate that SARS-CoV-2 will eventually attain endemicity and is likely to cause annual or biannual infection waves, similar to influenza and seasonal coronaviruses. Therefore, it is imperative to monitor the antigenic changes of SARS-CoV-2, as this information will be important in the selection of future vaccine strains.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mykytyn, Z. A., Rissmann, M., Kok, A., et al. (2022) Omicron BA.1 and BA.2 are antigenically distinct SARS-CoV-2 variants. bioRxiv. doi:10.1101/2022.02.23.481644. https://www.biorxiv.org/content/10.1101/2022.02.23.481644v1.
- Peer reviewed and published scientific report.
Mykytyn, Anna Z., Melanie Rissmann, Adinda Kok, Miruna E. Rosu, Debby Schipper, Tim I. Breugem, Petra B. van den Doel, et al. 2022. “Antigenic Cartography of SARS-CoV-2 Reveals That Omicron BA.1 and BA.2 Are Antigenically Distinct.” Science Immunology, June. https://doi.org/10.1126/sciimmunol.abq4450. https://www.science.org/doi/10.1126/sciimmunol.abq4450.