Introduction
Unwanted biomolecules need to be removed from serum, blood, urine, semen, spinal fluid, and other ex-vivo samples so that quantitative analysis of tiny molecules can be performed using LC-MS systems. The main components that need removing are proteins because these macromolecules become sticky and can damage chromatography columns. These large proteins can clog up the tiny holes in chromatography columns.
Chromatography
Chromatography is a standard separation technique used for quantitative analysis in life science studies. This technique can be carried out using either liquid or gas chromatographs, but both of these can become damaged by proteins. In addition to proteins, serum also contains other fatty acid derivatives called phospholipids that are also relatively sticky and can clog up analytical columns.
To resolve this problem, Porvair Sciences provide two types of 96-well filter plates that can be used to remove proteins from serum samples of up to 2ml in volume. The P3 or protein precipitation plate is the most simple and inexpensive method, while the C18 SPE plate is more advanced, but also more expensive. Which plate and method is chosen depends on the samples being used, the degree of sensitivity that is needed and the desired analyte.
Normally, samples that are prepared on P3 plates are appropriate for testing using standard LC-MS systems, while those processed on C18 SPE plates are more suited to highly sensitive QTOF and LC-MS analysis using capillary columns. If a mass spectrometer is used as a detection unit, the issue of phospholipid and protein contamination arises.
LC-MS
A common technique, LC-MS is often used to check for low levels of drug-like compounds and metabolites in blood or serum samples. At these low levels, the phospholipids pose a larger problem and one group of phospholipids called lysophospholipids, which are more complex to remove, can lead to inconsistent effects in the LC-MS signals detected.
Ion suppression is a phenomenon caused by lysophospholipids, where the high background signal induced by these substances masks the actual signal that is being produced by the analyte. Ion suppression can both increase and decrease the analyte signal making it difficult to correct for.
Protein Crash Technique
Although the protein crash method applied in the P3 plate fails to remove phospholipids and lysophospholipids, it is effective at eliminating proteins. This technique denatures the protein using acetonitrile or methanol. The protein “crashes” out of solution and turns into large, sticky globules which become bound to an open-pore structure filter. Underneath this filter is a smaller superhydrophobic filter which holds any liquids above it until a vacuum is used to push the liquid containing the desired analytes through the filter and finally into a collection plate positioned below.
Solid Phase Extraction
Solid phase extraction (SPE) is a traditional technique that uses a ‘sandwich’ of fine silica containing long-chain hydrocarbons between two supporting frits. This long carbon chain is oleophilic and slows the passage of globular proteins and fatty acids. As well as the loading step used in P3, the SPE plate requires additional conditioning and cleaning steps meaning this method takes more time and is more difficult to automate. On the other hand, the C18 silica is capable of removing nearly all the phospholipids from the samples. This article examines the suitability of different materials used to remove the phospholipids. Here, standard C18 Silica is compared with two different resins.
Assessing the C18 Silica Plate for Phospholipid Removal
For a device to work properly, it must allow the analytes to pass through easily, while retaining phospholipids on the sorbent. To evaluate this, the concentrations of analytes and phospholipids were determined using HPLC and colorimetric methods, respectively. Initially, it was proposed that a cation exchange mixed mode resin could be used as the sorbent for the following reasons:
- This would retain phospholipids via the positively-charged part of the hydrophobic tail and head group
- pH alteration would enable neutralization of the analytes to prevent retention through the ion exchange mechanism
- The methanol crash solvent would be enough to make sure that no retention occurs via hydrophobic interaction
Resin 1
A PCX resin, a mixed mode cation exchange resin, was used for the initial set of testing. A phosphatidylcholine standard was used to assess the removal of phospholipids. A traditional protein crash and a P3 hybrid with the PCX resin were then compared and neutral, acidic and basic methanol was used as the crash solvent.
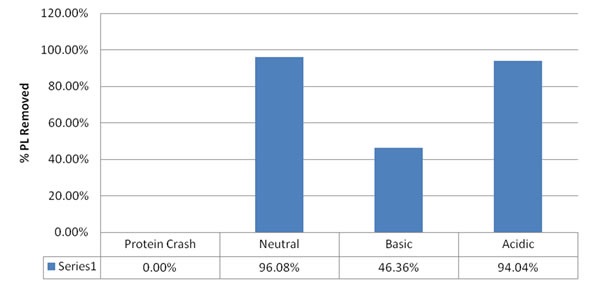
Figure 1. % removal of phospholipids with PCX resin and pH modified methanol.
As predicted, the results demonstrate that the traditional protein crash does not remove any of the phospholipids, whereas the PCX resin removes more than 90% of phospholipids when used with an acidic or neutral organic solvent. When a base-modified organic solvent is used, there is reduced retention of phospholipids.
The next experiment assessed the passage of analytes through the PCX sorbent un-retained. In a high organic eluent, no retention should occur through hydrophobic interaction and only the ion exchange mechanism could be a potential retention mechanism. There should be no interaction between sulfonate groups and acidic or neutral compounds, meaning no additional changes are required. Basic compounds will interact with sulfonate groups and would make it difficult to obtain good recovery. Therefore, testing was performed using the basic drug imipramine.
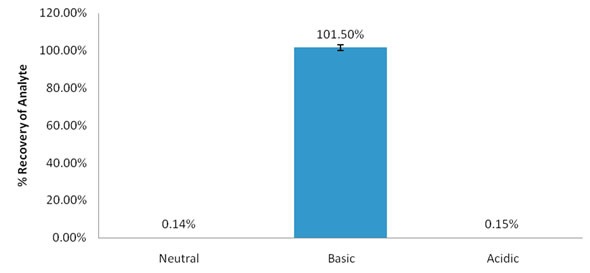
Figure 2. % recovery of imipramine with PCX and pH modified methanol.
The results demonstrate that imipramine is only recovered in a base-modified solution. This is the only condition where the analyte is in a neutral state and the ion exchange interaction is therefore prevented.
When these results are considered along with those from the first experiment, it becomes obvious that good retention of phospholipid cannot be achieved when base-modified organic solvents are used. On the other hand, this issue can be overcome by changing the solvent’s ionic strength so the pH is sufficient to shift the analyte to a neutral state while at the same time not preventing the retention of phospholipids.
Resin 2
Next, the PRP X400 resin was examined for retention of phospholipids using acidic, neutral and base modified organic solvents.
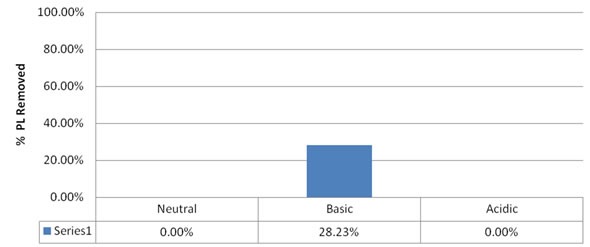
Figure 3. % removal of phospholipids with PRP X400 resin and pH modified methanol.
Given that the PRP resin is a divinylbenzene modified with sulfonate groups, it should be similar to the PCX resin. However, its performance with respect to phospholipids removal is extremely poor. Exactly why this difference occurs is not clear, but since previous work suggested cation exchange may not be a viable option, no additional work was carried out.
Summary
So far, the results demonstrate that good phospholipid removal can be achieved if a cation exchange resin is properly selected. However, significant method development would be needed to achieve acceptable analyte elution for basic compounds. It may be advisable to consider a simpler approach which relies on the long hydrophobic tail of the phospholipids strongly interacting with the selected resin. The interaction would probably not allow recovery in 100% organic solvents, but given that the procedure involves protein crash at a 3:1 ratio, it will work at about 75% organic solvent. This should be sufficient to allow the retention of phospholipids while also preventing the weaker interaction of analytes. It is possible that a C18 silica based sorbent may be adequate because without the need for additional steps, drying out of the sorbent bed is not an issue. Good retention of phospholipids can be achieved as long as the p3 frit is beneath the sorbent to enable initial wetting before the addition of plasma.
Initial Investigation of C18 for Phospholipid Removal
Experiment 1
In this experiment, 600µl of a 1mg/ml mixture of phosphatidylcholine in methanol was introduced into a well of a C18 Microlute plate. Then, 200µl of distilled water was added and the mixture was drawn through the C18 layer under vacuum using a Porvair Sciences’ vacuum manifold.
A Porvair Sciences’ Minivap nitrogen dry-down station was used to collect and evaporate the filtrate, which was then reconstituted in 600µl of chloroform in preparation for phospholipid testing. A control sample that was not passed through the C18 plate was also examined.
|
|
µg/ml
|
% Removed
|
|
Control
|
807.08
|
|
|
Sample 1
|
1.89
|
99.77%
|
|
Sample 2
|
5.40
|
99.33%
|
|
Sample 3
|
49.77
|
93.83%
|
|
Sample 4
|
91.89
|
88.61%
|
The outcome indicates that with a methanol to water ratio of 3:1, it is possible to remove about 90% of phospholipids. The experiment was again performed with acetonitrile instead of methanol and the outcome demonstrated that no phospholipids were retained. This is critical for choosing appropriate plate-based protocols for removing phospholipids.
Experiment 2 (Phospholipids Removal from Pig Plasma)
In the second experiment, six pig plasma samples were prepared by injecting 200µl of plasma into 600µl of methanol. These samples were then centrifuged to eliminate precipitated proteins and the supernatant was collected.
- Sample 1+2: No further preparation
- Sample 3+4: Filtered via 10mg C18 bed
- Sample 5+6: Filtered via a 30mg C18 bed
Using the Minivap, all samples were evaporated to dryness under nitrogen and reconstituted in 600µl of chloroform in preparation for phospholipid testing. For both C18 loadings, the concentration of phospholipids was reduced to below the limit of detection at 10µg/ml, which is a major reduction when compared to protein crash alone.
Experiment 3 (Analyte Recovery)
In the third experiment, three corticosteroids, namely prednisolone, corticosterone, and dexamethasone, were used to evaluate the recovery of analytes. Next, a solution of 20µg/ml of each corticosteroid was prepared in 75% water/methanol. Then 1ml of the analyte solution was filtered through a C18 bed and the filtrate collected and examined using HPLC.
Conclusion
All the experiments described above indicate that SPE using a C18 resin is a viable approach for removing phospholipids. While acetonitrile could not be used, a typical crash ratio of 3:1 methanol to plasma makes it possible to remove most of the phospholipids, while allowing most of the analytes to pass through easily. The study indicates that the combination of C18 sorbent and p3 frit may provide a suitable method for removing phospholipids, although more research is required to determine the elimination of lysophospholipds from these samples.
Acknowledgement
Produced from articles authored by Stephen Knight BSc. MA, Director of Sales & Marketing, Porvair Sciences Ltd, Wrexham, United Kingdom.
About Porvair Sciences
Porvair Sciences, specialists in the manufacture of microplate products, serve Life Sciences, Biotechnology, R&D and Molecular Biology with microplate solutions for all applications, from sample preparation to high throughput screening via our global distributor network.
Our range includes vacuum manifolds, sealers, evaporators and microtiter plates in all popular styles; deep well and shallow well storage plates, assay plates, luciferase reporter gene plates and liquid handling reagent reservoirs. We also provide custom microplate products for life science research. Our vacuum manifolds, essential for 96-well SPE sample preparation, are designed to work with most popular filter plates, including Waters, Millipore, Qiagen, Whatman, GE Healthcare, Varian, Biotage and, of course, Porvair.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.