Most pathogenic viruses, such as the causative agent of the current coronavirus disease 2019 (COVID-19) pandemic, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), affect normal human biology to co-opt cellular metabolism and replicate successfully. One key pathway is the suppression of innate host immunity.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Innate immune response
It was microbiologist Hans-Heinrich Hoffman who first called the host innate immune system-virus interaction a “molecular arms race,” with each side determined to gain the ascendancy.
Many researchers have suggested that the ability of a virus to cause disease could be in proportion to its ability to inhibit the early warning system of the host, the innate immune system. By this means, it would be able to achieve high viral loads before effective immune responses kick in.
Such information could also provide clues as to the potential for a virus to cause a pandemic. In the current outbreak scenario, the virus was found to be capable of causing a pandemic based on epidemiological observations of its spread and infection characteristics, over more than a month after its initial sequencing.
It should be possible to rapidly evaluate a novel virus for its pandemic potential based on the sequence alone, by comparing it to that of other members of the same family with known pathogenicity. It then becomes a mere matter of measuring how active its genes are, how its genetic pathways are likely to work, and the resultant disease-causing probability of the new virus.
Viral proteins and innate immunity inhibition
The current study aimed at developing a set of ‘high-content’ cell-based assays that can tell how capable a viral protein is of host innate immune inhibition. These can be used on a novel virus rapidly to assess pandemic potential.
Such screening assays have been used to identify drugs capable of disrupting cell pathways or operating states without knowing which proteins are affected.
The current effort makes use of both target-based screens to help understand the mechanism of inhibition of innate immunity components by individual viral genes. Another screen is independent of the mechanism, but examines the overall capacity of a viral gene to facilitate viral replication by inhibiting the immune response.
The flu-like symptoms of viral infections, in general, are due to the release of type I interferons (IFN), as shown by the occurrence of such symptoms in those treated with IFNs for hepatitis or multiple sclerosis. However, SARS-CoV-2 is capable of infecting humans and replicating to produce virions that can then spread to other humans without symptoms.
Pattern of antiviral innate immune response
Typically, the presence of viral nucleic acids in the cytoplasm triggers recognition by pattern-recognition receptors (PRRs), which initiates signaling pathways that lead to the shifting of Interferon Response Factor 3 (IRF-3) and Nuclear Factor κB (NFκB) to the nucleus.
These two transcription factors must be activated together to develop an antiviral response. Their activation in the nucleus leads to type I IFN expression along with that of other cytokines that alert the neighboring cells of viral attack, trigger adaptive immunity, and recruit specialized immune cells.
The type I IFN receptors activate their receptors on surrounding cells to lead to the translocation of Signal Transducer And Activator Of Transcription (STAT) transcription factors, sounding a general alarm against invasion.
Coronaviral genes inhibit key transcription factors
The researchers found that many different coronavirus genes inhibited the translocation of IRF-3 and/or NFkB to the nucleus. The strongest effect was found with NSP1, NSP3, NSP5, and Orf7a, but less so with NSP4, NSP10, NSP12, Env, and CaORF15.
Of these, CaORF15 and NSP9 have not been previously described to have such activity.
CaORF15 is a putative novel SARS-CoV-2 protein encoded within an alternate reading frame within the spike sequence. It is not found in SARS-CoV, but has been detected in the very closely related bat coronavirus RaTG13.
In fact, this sequence shares 98% identity with its homolog in the latter virus, compared to 93% for the spike sequence overall. This protein inhibits the translocation of IRF-3 to the nucleus, especially through the STING cellular pathway.
All coronaviral NSP5 proteins tested here inhibited IRF-3. Both IRF-3 and NFkB were suppressed by several different NSP10 and NSP12 genes, though only modestly.
However, the STAT1 pathway was mostly unaffected except by NSP1 and ORF6. These two genes have been reported to inhibit translation via different mechanisms.
SARS-CoV-2 genes show stronger innate immune suppression
SARS-CoV-2 genes produced much more powerful suppression of these transcription factors than the average effect produced by genes from the other viruses. This is due not only to the fact that this virus has more C-terminal ORFs, but also because these ORFs have intrinsically stronger effects compared to corresponding SARS-CoV genes.
These genes have no homologs in the other coronaviruses. Thus, SARS-CoV-2 is associated with the strongest suppression of the innate immune response activated by type I IFN.
Moreover, this virus contains NSP1, a powerful inhibitor of NFkB-activated gene expression.
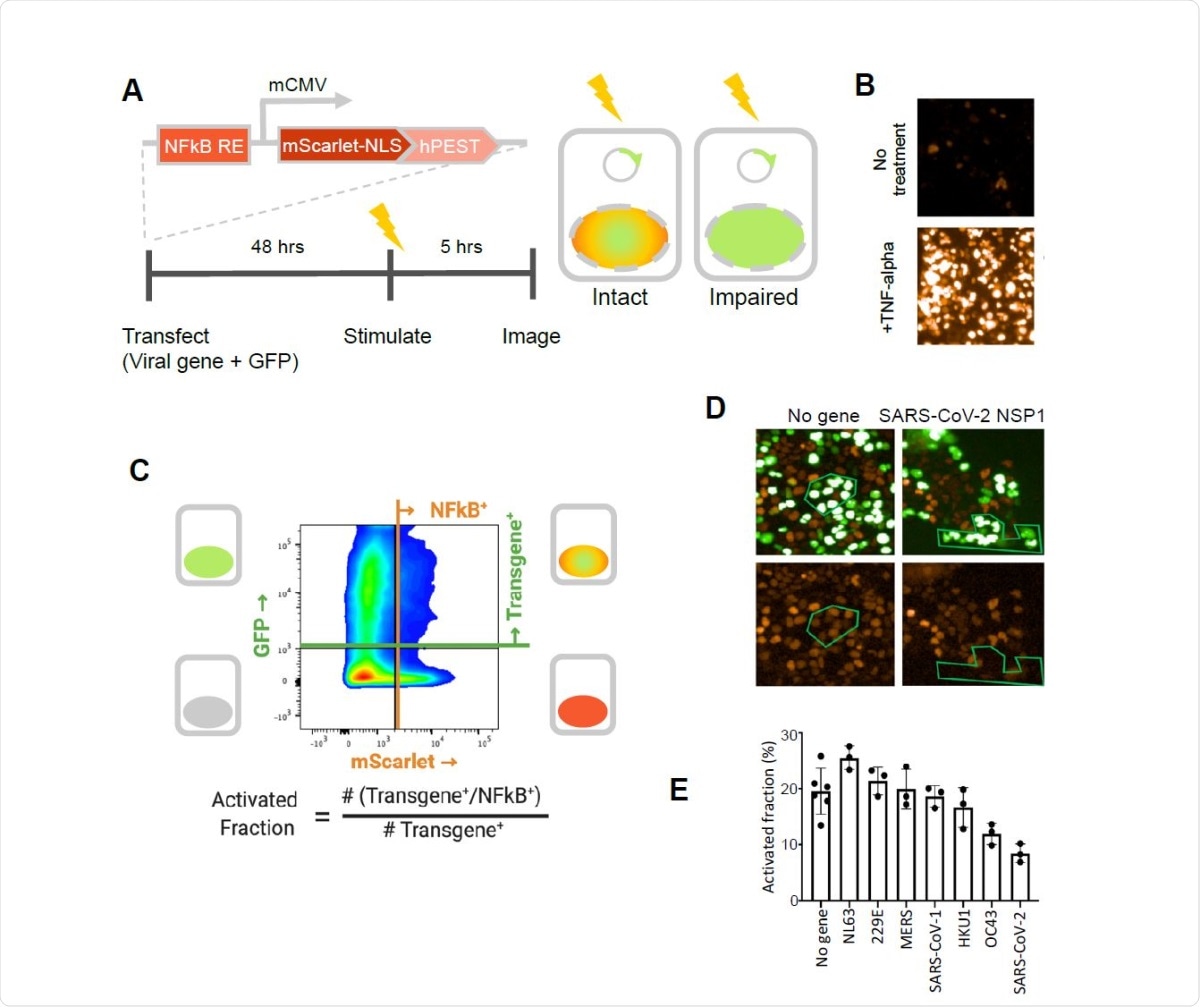
SARS-CoV-2 NSP1 strongly inhibits TNF-alpha activation of an NFkB reporter gene. A. A fluorescent NFkB reporter consisting of a 5x repeat of the NFkB consensus sequence (NFkB RE) upstream of a minimal CMV promoter (mCMV), and a human codon-optimized, nuclear localized mScarlet (mScarlet-NLS) fused to an hPEST degradation tag (hPEST) was constructed and stably integrated into HEK293 cells. Reporter HEK293T cells were co-transfected with expression plasmids encoding GFP and a virus gene; 48 hours later, 5 ng/ml TNF-? is added; and 5 hours later, red and green fluorescence was quantified. B. HEK293T cells stably transduced with the reporter responded to human TNF-? by expression of mScarlet. C. Flow cytometry analysis of transfected, TNF?-treated cells. Upper quadrants are transfected cells. The upper right quadrant represents double-positive cells that are transfected and also 338 express the inducible transgene. The activated fraction was calculated as the ratio of double positive cells to all GFP-positive cells. D. Reporter HEK293T cells were co-transfected with GFP and no-gene or SARS-CoV-2 NSP1 expression vectors, followed by TNF-? stimulation and imaging of red and green fluorescence. Green polygons highlight identical populations of cells in both images for representative comparisons. E. Activated fractions compared among NSP1s from several coronaviruses. Error bars represent the standard deviation of at least three technical replicates.
What are the implications?
This study shows the utility of a cell-based assay in reflecting the extent of viral replication in response to immune suppression.
Overall, SARS-CoV-2 proteins are strongly suppressive of the innate immune response compared to the average of all coronavirus genes put together, based on the following effects: IRF-3 and NFkB nuclear localization, NSP1 inhibition of NFkB-mediated gene expression, and effects in the yeast cell fusion assay.
In the latter, only the presence of active innate immune suppression yields active virus. Thus, the recovery of the virus indicates the presence of such viral proteins, in general, but if the host inhibition is very powerful, the virus may not be able to replicate at all. This appears to be the case with SARS-CoV-2 NSP1.
Importantly, the presence of the CaORF15 gene within the spike sequence means that it is part of many vaccines based on the wildtype spike, and as such, may hinder the generation of immunity in response to the vaccine. This is not the case with the Pfizer/BioNTech vaccine, which has a modification within this region.
The suppression of innate immune responses is more likely to be aligned with asymptomatic spread rather than increased disease severity.
This suite of pathway-based and mechanism-agnostic assays could serve as the basis for rapid in vitro prediction of the pathogenicity of novel viruses based on provision of sequence information alone.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.