Caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) agent, the coronavirus disease 2019 (COVID-19) pandemic continues to spread across the globe. It has infected over 50 million, taken over 1.26 million lives, and has severely impacted many national economies. These alarming developments have triggered a wave of intensive research into how the SARS-CoV-2 agent infects and replicates within host cells. A better understanding of the behavior of the SARS-CoV-2 pathogen within host cells can aid in developing therapeutic methods that mitigate its potentially lethal effect on those infected.
A recent study by researchers at Washington University School of Medicine, Saint Louis, and published on the preprint server bioRxiv* in October 2020 shows what the protein translation process looks like in specific terms. In doing so, the study has provided new insights that might prove helpful in developing new therapies for COVID-19 patients. “These data reveal the key role of mRNA translation in SARS-CoV-2 replication and highlight unique mechanisms for therapeutic development,” say the researchers.
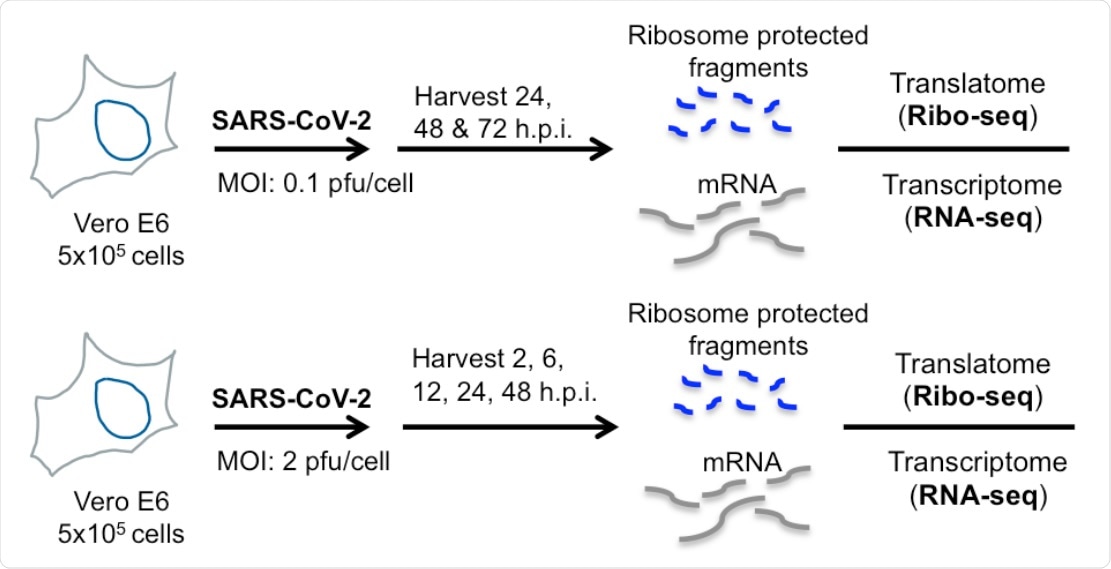
Ribo-seq reveals uneven ribosome occupancy on SARS-CoV-2 RNAs during low MOI infection. (A) Schematic diagram of Ribo-seq and RNA-seq experiments conducted in this study. Vero E6 cells were infected at 0.1 pfu/cell and cells were processed for RNA-seq and Ribo-seq at 24, 48 and 72 hpi.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Structural elements that regulate viral transcription
The study profiles the ribosome occupancy of various viral and host messenger RNAs (mRNAs). It thus shows how the virus quickly hijacks the cell’s transcriptional pathways, with a majority of cellular mRNA and a significant percentage of ribosome-associated mRNAs being virally derived at 24 and 48 hours post-infection.
SARS-CoV-2 uses host translational machinery and also regulates mRNA translation by the post-transcriptional suppression of host genes to disarm the host immune system and allow high levels of viral replication.
The viral NSP1 both induces cleavage of the host mRNA and inhibits the initiation of their translation. This may allow the virus to overcome the antiviral type I interferon response in infected cells – and this has been shown to occur in SARS-CoV-2. Thus, the transcriptome-based earlier studies may not reflect the virus’s capacity to modulate host immunity.
The researchers find a temporal and abundance-linked pattern of viral mRNA occupancy by ribosomes. The N-protein was translated in abundance, but thereafter there was a steep drop in the number of ribosomes translating the ORF10 coding sequence. The translation start sites of the N- and M-ORFs showed peak ribosome occupancy. With only a low level of mRNA for ORF1ab, this polyprotein appeared to be translated with great efficiency.
Ribosome occupancy was especially and persistently high in the ORF6/ORF7 region. This is predicted by computational modeling to have a hairpin loop structure, which may cause the ribosomes to pause during the translation of the genomic RNA. This may be the reason for the high occupancy that was observed. The function of this site is not clear; it may be just a pause site or may regulate the proportion to which ORF6 and ORF7 are translated – or it could have another translational function altogether.
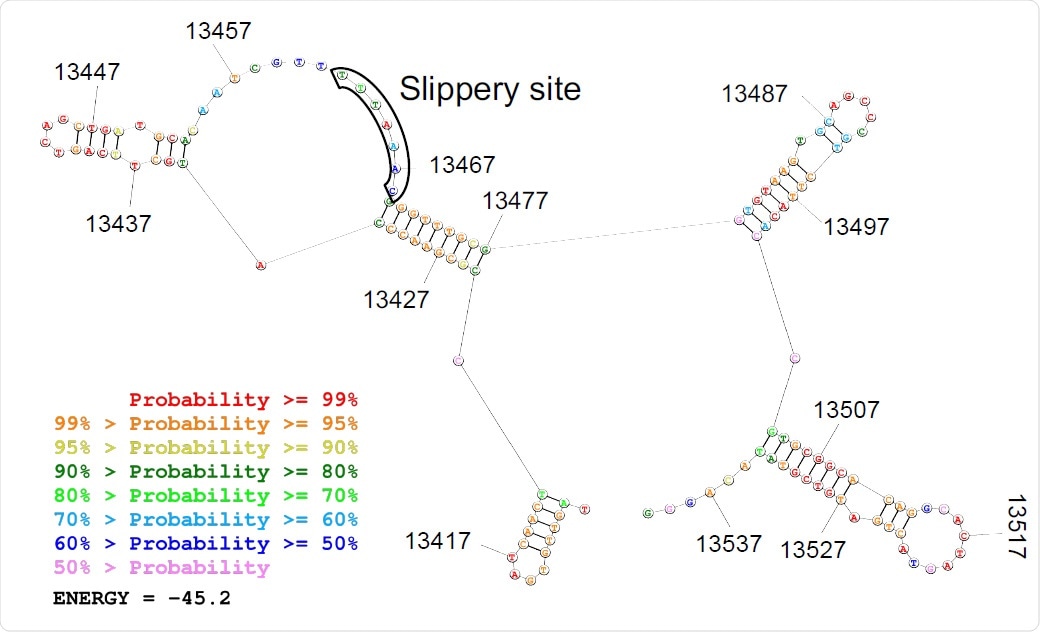
Predicted secondary structure of the frameshifting site is depicted.
Efficient ribosomal frameshifting strategy
This RNA virus has a long genome, about 30 kb, of which about two-thirds is made up of an open reading frame, ORF1ab. This translates a polyprotein, which is then split into several viral non-structural proteins (NSPs) essential for viral replication. The successful translation of the polyprotein depends on a programmed -1 ribosomal frameshift (PRF) that occurs at the overlap of the two ORFs, ORF1a and ORF1b. The PRF requires a slippery sequence of seven nucleotides and an RNA pseudoknot structure that has also been preserved. Both of these are highly conserved among coronaviruses.
The pseudoknot causes the ribosomes engaged in protein synthesis to stop on the slippery sequence. This is thought to make the frameshifting event more efficient, compared to the alternate stem-loop structure that is found in retroviruses that achieve only 5-10% efficiency. Once the -1 slippage is achieved, the pseudoknot unravels and the translation of ORF1b proceeds to the end. However, no empirical, as opposed to theoretical, evidence exists for the varying behavior of ribosomes within the frameshifting element (FSE) for either type of virus.
Again, ribosomes on the FSE were relatively few throughout the infection, indicating a highly efficient PRF. Upstream of this site, too, ribosome occupancy was low, either due to steric hindrance or perhaps due to the presence of an alternative RNA structure in this region.
Immune responses disarmed by translation block
In addition, the mRNAs for many inflammatory cytokines were upregulated, but these proteins themselves were not translated in correspondingly high amounts. While some cytokines such as EGR1, EGR3 and IFIH1 were exempted from this viral suppression, immune response genes showed broad translational repression. This included the interferon-related inflammatory cytokines IL2 and I6, but except for the interleukins IL11 and IL1A. This pattern was confirmed in viral cultures of airway epithelial cell lines, indicating their physiological relevance.
Alternative translation initiation events
On the other hand, the host cell appears to respond by using alternative sites to initiate translation, thus switching to different host responses and using a varied repertoire of immune responses. The alternative translation start sites within several viral genes, including those encoding the spike, membrane and nucleoprotein proteins, may cause the production of isoforms or shortened forms of these viral antigens, which may have important roles to play.
A host of cellular genes with unknown functions were also found to be translated at higher levels in infected cells.
Translation block regulates host responses
Overall, the researchers found that viral mRNAs are not translated more efficiently than host mRNAs, and the high abundance of the former is therefore the reason for the relatively higher amounts of viral proteins. A possible exception is the somewhat greater efficiency of translation of the S, E and ORF1ab mRNAs.
The cytokine storm seen in severe COVID-19 patients is difficult to explain in view of the dampened immune responses produced by SARS-CoV-2 infection. However, this may be the result of immune cell infection, which are less readily infected but respond to infection by secreting molecules that regulate immune responses.
Conclusion
The researchers conclude, “Our finding that induction of inflammatory and innate immune responses can be limited at the level of mRNA translation provides a paradigm-shifting mechanism of how SARS-CoV-2 can encounter immune responses.”
The study could lead to the development of therapeutics that target viral RNA and modulatory proteins that target viral mRNA translation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Puray-Chavez, M. et al. (2020). The Translational Landscape Of SARS-Cov-2 And Infected Cells. bioRxiv preprint. doi: https://doi.org/10.1101/2020.11.03.367516, https://www.biorxiv.org/content/10.1101/2020.11.03.367516v1
- Peer reviewed and published scientific report.
Puray-Chavez, Maritza, Nakyung Lee, Kasyap Tenneti, Yiqing Wang, Hung R. Vuong, Yating Liu, Amjad Horani, et al. 2022. “The Translational Landscape of SARS-CoV-2-Infected Cells Reveals Suppression of Innate Immune Genes.” Edited by Kellie Jurado. MBio 13 (3). https://doi.org/10.1128/mbio.00815-22. https://journals.asm.org/doi/10.1128/mbio.00815-22.