Single-cell cloning involves generating a cell line from a single initial cell isolated from a heterogeneous population, typically obtained through transfections, transductions, or primary cells derived from tissue or biopsy samples.1
With the emergence of novel methodologies, the life science drug development field has expanded beyond small molecules to encompass more targeted biologics and advanced therapy medicinal products (ATMPs).
Within this expanding landscape, single-cell workflows have played a pivotal role, offering significant clinical and scientific implications across various interdisciplinary applications, such as multi-omics analysis,2 immune-oncology,3 rare cell identification,4 drug discovery,5 stem cells (in regenerative medicine),4,6,7 biologics,8 organoids,9,10 and epigenetics.11,12
In recent years, the development of cell lines for analytical, bioproduction, and therapeutic purposes has become a critical workflow, with monoclonality being a crucial requirement (Figure 1).
Pursuing more targeted therapies for diverse disease conditions has spurred advancements in single-cell workflows, particularly in cloning, which has become an indispensable necessity in biopharmaceutical research and product development.
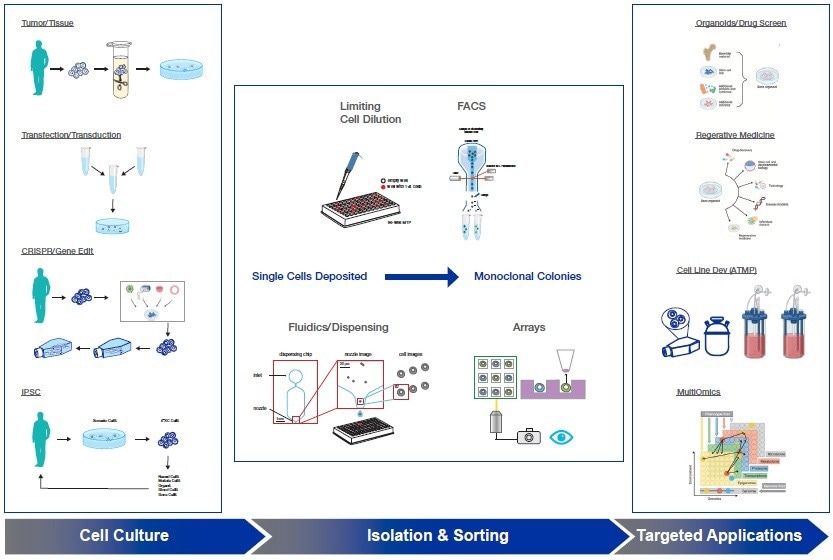
Figure 1: Single-Cell Workflow and Applications. Image Credit: Cell Microsystems
Current methods
Limiting dilution and single-cell dispensing
Two commonly utilized techniques for isolating single cells are limiting dilution and single-cell dispensing, occasionally assisted by fluorescence cell sorting.
Though widely employed, these techniques are burdensome, time-consuming, and possess notable drawbacks. They demand meticulous sample preparation, extensive training, and equipment maintenance and often entail multiple rounds of cloning to generate a monoclonal cell line.13
Consequently, these disadvantages frequently result in diminished cell viability and proliferation. Despite these limitations, limiting dilution is the most convenient and cost-effective method for single-cell isolation.14,15
This approach involves generating a monoclonal cell line from a heterogeneous pool through sequential dilutions, followed by individual clone analysis using image-based techniques and subsequent expansion of selected clones.
Although retrieving individual cells from diluted cell suspensions may appear straightforward, employing manual pipetting or automated robotic platforms often proves challenging when obtaining an adequate number of desired monoclonal clones.
It is common to encounter wells with multiple cells or even the absence of cells, leading to a shortage of viable clones. The inability to ascertain whether cells were genuinely isolated before expansion introduces uncertainty regarding the origin of subsequent colonies.16
Flow cytometry and cell sorting
Despite creating an illusion of improvement over limiting dilution, utilizing flow cytometry and cell sorting presents challenges.14,17 In this technique, cells are examined and sorted based on the hydrodynamic focusing phenomenon and specific cellular characteristics.
Fluorescence-activated cell sorting (FACS) separates a single cell from a cell suspension with a certain degree of purity. The cell sorting instrument can accurately and efficiently place a single cell into a microplate well.
Benchtop cell sorting systems equipped with multiple lasers and colors enhance the precision and speed of the sorting and selection process.18 However, these advancements necessitate significant gating requirements and setup for each population to be sorted, ensuring the retrieval of the maximum number of viable clones.19
In some instances, the availability of fluorescently tagged antibodies targeting surface epitopes can pose a highly complex reagent preparation process. Cell sorting and dispensing can damage cells, altering their metabolic state and inducing oxidative stress.20
It is common for only 20% of isolated single cells to produce usable colonies due to a fraction of FACS-sorted cells being dead or experiencing hindered growth.19,21
Cells subjected to FACS sorting exhibit a 50% increase in reactive oxygen species, indicating that their metabolism shifts from an anabolic state to a catabolic state as they transition from proliferating in cell culture to the stressful environment of sorting and dispensing.
This metabolic transition reduces reductive biosynthetic reactions involving NADPH. Cells undergoing this transition activate apoptotic genes or halt S-phase synthesis. Consequently, diminished outgrowth is frequently observed in single-cell progenitors initiated through FACS, and achieving the necessary outgrowth yields often takes longer.20
Despite widespread use in academic and biopharmaceutical settings, these techniques have significant drawbacks. Firstly, they require the cell preparation to be a single-cell suspension, rendering them unsuitable for adherent cells without extensive enzymatic or physical treatment.
Secondly, both methods rely on statistical probabilities to assert monoclonality, necessitating multiple rounds of serial cloning to establish a clonal cell line, and often overestimate the likelihood of monoclonality due to the presence of cell clusters.21
Consequently, verifying a single-cell progenitor entails the laborious and time-consuming microscopic examination of all microplate wells, lacking a documented image of a single cell. Therefore, demonstrating monoclonality with these techniques is arduous and challenging.
The systems employed for these methods of single-cell clone generation comprise multiple platforms, components, and steps.
This can be economically burdensome, restrict compatibility and availability, and frequently necessitate troubleshooting at various workflow stages, further adding to the complexity of this already demanding process. (Table 1)
Table 1. Cell Sorting and Isolation Methods. Source: Cell Microsystems
| Isolation Methods |
Description |
Advantages |
Disadvantages |
Cost |
Limiting Dilution -
Manual |
Serial dilution until solution is statistically calculated to be one cell per microliter |
Established, simple and familiar protocols; perceived to be low cost |
High failure rate; error-prone; tedious; extended experimental timeline (~10 weeks); additional equipment/space required (biosafety cabinet, incubator, cell counter); contamination risk; high risk of isolating multiple cells; manipulation of cells can perturb expression profiles; not designed for bulk sorting |
$ |
Limiting Dilution -
Automated |
Robotic-controlled micropipettes |
High accuracy; fluorescence can be used |
Lack of software analysis increases time and effort to get reasonable results for isolation & selection. |
$$ |
| Flow Sorting |
Microdroplets with single cells are isolated by electric charge at high pressure |
Enables bulk or single-cell sorting high accuracy and precision for identifying cells/populations of interest; fluorescent markers can be used to isolate sub-populations. |
Requires separate single-cell dispenser; low yield; requires off-platform propagation for cell line development; not amenable to organoid/3D biology; equipment and manual labor- requires hands-on attention; fluidics perturb cell metabolism; perturb expression profiles and damage cells |
$$ |
| Microfluidic platforms |
Microfluidic chips isolate single cells in flow channels |
High-throughput; reactions can be performed on-chip; reduced reagent costs |
High failure rate; prone to contamination; highly complicated fluid mechanics can complicate outcomes; lack of imaging options |
$$ |
| Cell Dispensing (droplet) |
Single-cell trapped in microfluidic drops |
Single cells can be imaged in a flow path |
Highly complicated fluid mechanics can complicate outcomes |
$$ |
| Cell Raft Technology |
Single cells grown in a specialized culture dish; one single platform for integrated imaging, analysis, isolation. |
Able to isolate up to 400 clones from each array into 96-well plates, with each plate giving rise to >90% single-cell growth into colonies; can propagate stem cells/iPSCs, organoids, or screen T-cells; fast, ease of tracking and tracing of clonal propagation unique, powerful software drives selection and isolation of cells based on particular end-user requirements |
Not ideal for high-throughput single-cell genomics; not designed for bulk sorting; cannot be integrated with sample prep methodologies. |
$ |
| Optofluidic Technology |
Uses light and millions of light-actuated pixels to move individual cells so they can be isolated, cultured, assayed, and exported |
Integrated workflow |
Limited number of cells; chip only has 5,000 positions; not every nanopen position is occupied; very limited applications; requires a fully dedicated lab technician to operate; technology has not been fully adopted yet |
$$$$$$ |
Cell dispensers
In recent times, the market has witnessed the emergence of several new products dedicated to cell sorting and dispensing, each promoting their system as a means to streamline the generation of single-cell clones. These setups exhibit a wide range of specifications and costs, varying from $150,000 to millions of dollars.
Among these systems, cell dispensers are the most prevalent. These platforms employ microfluidics in combination with bright field imaging or fluorescence detection to deposit a single cell into a well of a 96- or 384-well collection plate.
They aim to enhance the limitations of limiting dilution techniques by directly acquiring an isolated single cell, potentially improving confidence in monoclonality. However, these systems offer limited benefits.
The subsequent outgrowth of the isolated single cells necessitates separate procedures, requiring additional equipment, resources, and space, which incurs additional costs beyond the initial platform investment, which can be substantial, ranging from several hundreds of thousands to millions of dollars.
Moreover, the physical manipulation involved in fluidic channels and the impact of droplet dispensing can potentially harm the isolated cell, leading to perturbations in expression profiles and reduced outgrowth (Figure 2).22
These platforms are specialized in their function and do not support other forms of selection or propagation. Consequently, researchers seeking single-cell workflows to propagate stem cells/iPSCs, organoids, or T-cell screening cannot benefit from these systems.
Although single-cell dispensers represent an improvement over traditional manual methods, they do not fully fulfill the requirements for facilitating clones’ rapid and efficient development.
With a saturated market offering a variety of platforms, researchers are faced with the challenge of navigating and selecting the most suitable and cost-effective system for their specific needs.
While innovative techniques like microfluidic platforms and automated clone pickers show promise for screening, selecting, and isolating monoclonal clones, they still fail to deliver a high volume of viable monoclonal colonies without imposing resource constraints.
There is a clear demand for more comprehensive tools encompassing the entire workflow, from single-cell separation to outgrowth, while remaining applicable across multiple cell lines and types.
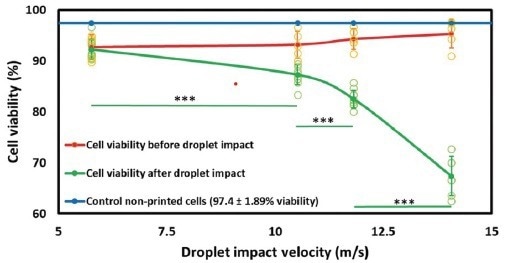
Figure 2. Droplet Impact Can Damage Cell Viability. (adapted from Ng et al., 2022). Image Credit: Cell Microsystems
Overcoming the challenges
In response to the needs of researchers and the shortcomings of existing methods (Table 2), there is a clear demand for a system or technology that enables the isolation of single cells while maintaining natural cell cycle kinetics and preserving biochemical equilibrium.
To address these requirements, innovative technologies have been developed, such as the CellRaft® Technology (Figure 3), which offers flask-like culture conditions at the resolution of a single cell and employs gentle and automated isolation based on image-based attributes related to function, gene expression, and morphology.
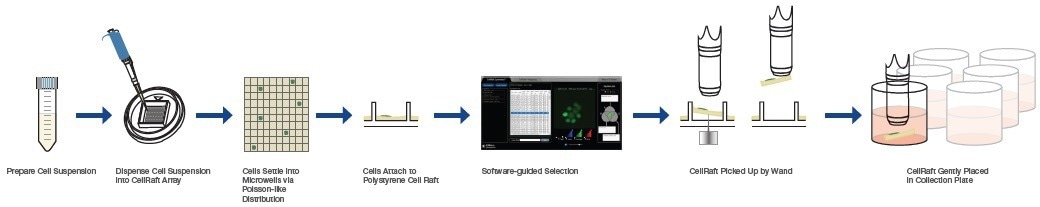
Figure 3. CellRaft Technology Combines the Power of Flask-like Culture Conditions and Single-Cell Separation to Produce High Viability Cells, Colonies, and Organoids. Image Credit: Cell Microsystems
Table 2. Comparison of the different technologies from the perspective of an experimental workflow. Source: Cell Microsystems
| Experimental Needs |
Limited Dilution (manual) |
Limited Dilution (automated) |
Flow Sorting |
Cell Dispensing (droplet) |
OptofluidicTechnology |
CellRaft Technology |
| Main Applications |
Single Cell Cloning |
Single Cell Cloning |
Single Cell Isolation |
Single Cell-Omics; Cell Line Development; Gene Therapy |
Clonal Cell Line Development |
Cell Line Development Single Cell cloning; iPSC & Organoids Engineering, Development and Maturation |
| Throughput (number of cells for single cell propagation) |
Low |
High |
High |
High |
High |
Medium |
| Cell Viability |
Very Low |
Very Low |
Low |
Low |
High |
Very High |
| Outgrowth Efficiency |
Negligible |
Very Low |
Medium |
Medium |
Low |
Very High |
| Visual Control |
None |
None |
None |
Partial |
Yes |
Yes |
| Cell Selection |
None |
None |
Limited |
Limited |
Limited |
Yes fully capable |
| Starting Number of Cells Needed |
High |
High |
Moderate |
Moderate |
Low |
Moderate |
| Flexibility (Own Protocols) |
Yes |
Yes |
No |
No |
No |
Yes |
| Software |
None |
Used for Robotic Control |
Analyze, Segregate |
Analyze, Segregate |
Analyze, Robotic Control |
Analyze, Isolation, Monitoring as a Function of Time |
| Lab Skills Needed |
Low |
Low |
High |
Low |
Very High |
Low |
| Integration with Lab Management System |
No |
Yes |
Yes |
No |
Yes |
no |
| Integrated Workflow |
No |
No |
No |
No |
Yes |
Yes |
| Footprint in the Lab |
Negligible |
Large |
Large |
Small |
Very Large |
Small |
| Number of Cell Types Demonstrated |
NA |
~10-25 |
>100 |
<20 |
2 to 1 |
~100 |
| 2D or 3D Biology |
No |
No |
No |
No |
No |
Yes |
| Robotic Compatible |
None |
Yes |
Some |
No |
Yes |
No |
| Real Time Live Cell Image Analysis |
No |
No |
No |
Yes |
No |
Yes |
| Track and Trace (Time Course/Audit Trail) |
No |
No |
Limited |
Limited |
Yes |
Yes |
| Image versus Signal |
None |
Image |
Signal |
Image |
Image |
Image |
| Proof of Monoclonality |
Indirect |
Indirect |
Indirect |
Indirect |
Direct |
Direct |
The CellRaft AIR® System23 represents the implementation of this technology as an integrated platform for the growth, scanning, analysis, and isolation of single-cell derived monoclonal colonies.
Central to this system is the CellRaft® Array, a cell culture dish consisting of 10,000-150,000 microwells known as CellRafts. These microwells enable the gentle settling of cells by gravity, resulting in their distribution across the array in various single, double, or clustered combinations.
Utilizing the Poisson distribution model, approximately 40%-60% of CellRafts are populated by single cells, depending on the seeding density. The unique design of the CellRaft Array allows cells to share the same media and extracellular growth factors or cytokines, mimicking the conditions of flask or reactor cultures.
Consequently, the physical isolation of single cells is achieved without perturbation, ensuring the absence of biochemical or physiological changes. The CellRaft AIR System offers capabilities for individual imaging and analysis of cells using bright field or fluorescent imaging modalities.
The accompanying CellRaft Cytometry™ software facilitates the selection of clones that meet the specific attributes defined by the user or application.
The system automates the acquisition, isolation, and retrieval of monoclonal colonies through the mechanical actuation of a magnetic wand, which gently transfers the colony-containing CellRaft to a 96-well collection plate.
The CellRaft AIR System can handle imaging, tracking, analysis, and automated isolation of colonies from single cells within a single instrument, without any minimum sample size requirement.
The absence of microfluidic separation or perturbation in the CellRaft Array ensures the viability and vitality of single cells, allowing them to develop into healthy clones.
By promoting cell-to-cell communication during clonal development, the shared media across the array fosters the generation of 10X to 50X more viable and highly proliferative monoclonal colonies than other systems.
This improvement is significant as it helps select cells not resting, undergoing early apoptosis, senescence, or other conditions that hinder or prevent their propagation after isolation.19
The CellRaft AIR System exhibits remarkable versatility, enabling colony growth from primary, adherent, or suspension cells, including iPSCs and immune cells. Users can cultivate iPSCs into 3D cell systems, such as organoids, for applications spanning cancer immunology, multi-omics, and cellular heterogeneity studies.
At the heart of the CellRaft AIR System lies the CellRaft Cytometry software, which serves as its "brains." This software facilitates the selection, scanning, and imaging of thousands of CellRafts, enabling the automated identification and isolation of desired clones. (Figure 4).
With real-time interaction capabilities, users can seamlessly navigate through the software's many features, which integrate smoothly with the hardware.
The software can be used on and off the system, providing flexibility for cellular data analysis on desktop or laptop computers. Key features of the CellRaft Cytometry software include:
- Versatility – multiparameter analysis (time, morphology, phenotype)
- Automated CellRaft identification and isolation, resulting in software-guided biology decisions
- Unbiased CellRaft selection, reducing errors in identification
- Easy template creation with a QuickStart library
- Savable user-defined parameters for assay accuracy and consistency
- Track and trace capability for an audit trail
- Simultaneous scanning and data analysis in real-time.
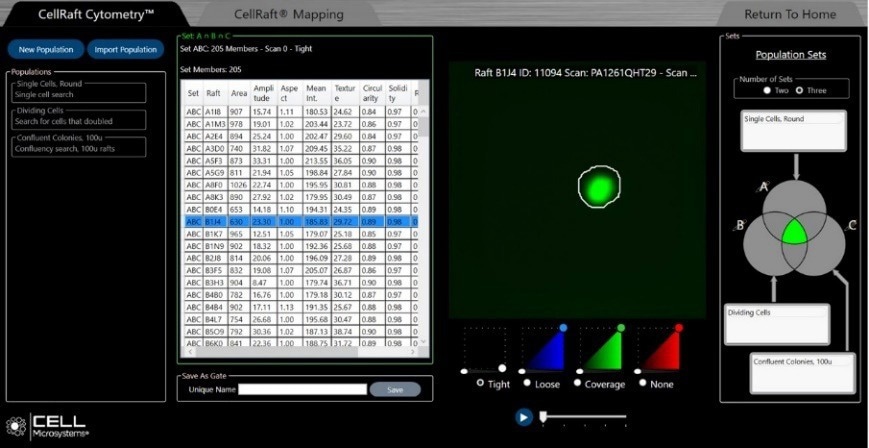
Figure 4. A single cell on a CellRaft can be readily identified using CellRaft Cytometry (green contour defines the boundaries of the area of interest). The Venn diagram shows the characteristics defined by the user to identify the cells of interest for isolation. The table identifies the contents of each CellRaft. Image Credit: Cell Microsystems
Workflow
In the CellRaft AIR System workflow, cells are plated on the CellRaft Array and loaded into the system platform. The cells are imaged using three-channel fluorescence and bright field microscopy, and sorting is performed based on user-defined thresholds, filtering, and gating (Figure 4).
The software analyzes expression, time, and morphology, and automatically isolates the desired CellRaft according to user-defined criteria.24 The system can isolate individual, undisturbed cells or colonies in a full 96-well plate in under an hour for expansion and downstream analysis.25
Compared to other available systems, the CellRaft AIR System offers several advantages. It streamlines the single-cell workflow by integrating imaging, identification, and isolation within a single instrument and consumable.
The system achieves high clonal yields with robust viability and cell proliferation, surpassing other platforms that require additional space and instruments.
Automating all steps provides real-time imaging, identification, and isolation of cells and small colonies for outgrowth in 96-well plates (Figure 3). This accelerates the timeline for obtaining results and reduces costs and contamination risks.
The system includes several onboard assays for cell characterization, co-culturing, cell-drug interactions, and cell-cell interactions.25 Importantly, the CellRaft AIR System supports a wide range of cell types and 3D cell systems, including stem cells (such as iPSCs), animal or human cells, primary cells, immortalized cells, adherent cells, suspension cells, and organoids.25
Recent experimental and cost analyses comparing the CellRaft Technology to limiting dilution demonstrate the superior outgrowth efficiency, time savings, and cost-effectiveness of the CellRaft Technology (Table 3).
CellRaft AIR System has been successfully used for single-cell workflows with approximately 100 cell lines and types (Figure 5). The development and validation of new workflows continue to expand its capabilities.

Figure 5. Timeline of Raji cell colony growth as an example of direct proof of track and trace functions. Image Credit: Cell Microsystems
Table 3. Side by side comparison to determine the savings in plastic, reagent, and media cost during the production of either 100 or 500 clones from Limiting Dilution versus CellRaft Technology. *Outgrowth efficiency measures the number of colonies obtained from single-cell deposits. Source: Cell Microsystems
| Economic Advantage of Cell Raft Technology versus Limiting dilution |
| |
|
Limiting Dilution |
CellRaft Technology |
| |
|
|
100 Clones |
500 Clones |
|
100 Clones |
500 Clones |
| Cell Line Category |
Representative Examples |
Outgrowth Efficiency* |
Total Cost-USD |
Hands on time (hours) |
Total Cost-USD |
Hours |
Out-growth Efficien-cy* |
Total Cost-USD |
Hands on time (hours) |
Total Cost-USD |
Hands on time (hours) |
| Production Cell Lines |
CHOK1 ; HEK293 |
9.4% to 30% |
$150-$446 |
4 to 12 |
$670-$2086 |
18 to 56 |
96% |
$75 |
0.6 |
$225 |
1.2 |
| Standard Cell Lines |
HeLa; C2C12; VERO |
2.9% to 22% |
$187-$1341 |
5 to 36 |
$894-$6075 |
24 to 180 |
66 to 90% |
$75 |
0.6 |
$224-$299 |
1.2-1.6 |
| Cancer Cell Lines |
HT-1080; HT-29; K562; C6 |
4.7% to 30% |
$150-$900 |
4 to 23 |
$670-$4134 |
18 to 111 |
75 to 100% |
$75 |
0.6 |
$224-$261 |
1.2-1.4 |
| Stem Cell Lines |
KYOU |
5% |
$1480 |
21 |
$7409 |
105 |
96% |
$150 |
0.6 |
$433 |
1.2 |
Conclusions
The traditional methods of limiting dilution and single-cell sorting have long been used to develop monoclonal cell lines but have been associated with several significant drawbacks. These methods often result in cellular damage, slow workflows, and uncertainties regarding clonality.
While newer single-cell dispensers have improved clonality assurance and confluence, they can still lead to low yields due to perturbation caused by the selection process.
There is an increasing need for a wider range of selection capabilities, particularly for stem cells/iPSCs, organoids, adherent cells, and rare cell types. The CellRaft Air System offers a comprehensive solution by integrating imaging, tracking, analysis, and automated isolation of verified monoclonal cultures.
It provides notable advantages over other systems, eliminating the need for additional space, equipment, and manual labor while ensuring cost-saving, viability, and high outgrowth rates.
The versatility of the CellRaft Air System makes it suitable for numerous applications, including cell line development, CRISPR gene editing, stem cell culture, organoid development, and single-cell genomics. Its broad range of capabilities makes it an ideal platform for various fields.
Ultimately, the CellRaft Air System has the potential to streamline and expedite the drug development process, leading to more rapid delivery of new drugs to the market.
References and further reading
- Kline, C. The art of generating single cell clones. BiteSize Bio https://bitesizebio.com/42870/to-be-a-clone-or-multiclonal/ (2019).
- Mincarelli, L., Lister, A., Lipscombe, J. & Macaulay, I. C. Defining Cell Identity with Single-Cell Omics. Proteomics 18, 1700312 (2018).
- Gohil, S. H., Iorgulescu, J. B., Braun, D. A., Keskin, D. B. & Livak, K. J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nature Reviews Clinical Oncology 2020 18:4 18, 244–256 (2020).
- Gach, P. C., Attayek, P. J., Herrera, G., Yeh, J. J. & Allbritton, N. L. Isolation and in vitro culture of rare cancer stem cells from patient-derived xenografts of pancreatic ductal adenocarcinoma. Anal Chem 85, 7271–7278 (2013).
- Heath, J. R., Ribas, A. & Mischel, P. S. Single-cell analysis tools for drug discovery and development. Nature Reviews Drug Discovery 2015 15:3 15, 204–216 (2015).
- Zheng, H. et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 68, 127–140 (2018).
- Kester, L. & van Oudenaarden, A. Single-Cell Transcriptomics Meets Lineage Tracing. Cell Stem Cell 23, 166–179 (2018).
- Diep, J. et al. Microfluidic chip-based single-cell cloning to accelerate biologic production timelines. Biotechnol Prog 37, e3192 (2021).
- Brazovskaja, A., Treutlein, B. & Camp, J. G. High-throughput single-cell transcriptomics on organoids. Curr Opin Biotechnol 55, 167–171 (2019).
- Youk, J., Kwon, H. W., Kim, R. & Ju, Y. S. Dissecting single-cell genomes through the clonal organoid technique. Experimental & Molecular Medicine 2021 53:10 53, 1503–1511 (2021).
- Tedesco, M. et al. Chromatin Velocity reveals epigenetic dynamics by single-cell profiling of heterochromatin and euchromatin. Nature Biotechnology 2021 40:2 40, 235–244 (2021).
- Meir, Z., Mukamel, Z., Chomsky, E., Lifshitz, A. & Tanay, A. Single-cell analysis of clonal maintenance of transcriptional and epigenetic states in cancer cells. Nature Genetics 2020 52:7 52, 709–718 (2020).
- Riba, J., Schoendube, J., Zimmermann, S., Koltay, P. & Zengerle, R. Single-cell dispensing and ‘real-time’ cell classification using convolutional neural networks for higher efficiency in single-cell cloning. Scientific Reports 2020 10:1 10, 1–9 (2020).
- Ye, M., Wilhelm, M., Gentschev, I. & Szalay, A. A Modified Limiting Dilution Method for Monoclonal Stable Cell Line Selection Using a Real-Time Fluorescence Imaging System: A Practical Workflow and Advanced Applications. Methods Protoc 4, 1–14 (2021).
- Lefkovits, I. & Waldmann, H. Limiting dilution analysis of the cells of immune system I. The clonal basis of the immune response. Immunol Today 5, 265–268 (1984).
- Evans, K. et al. Assurance of monoclonality in one round of cloning through cell sorting for single cell deposition coupled with high resolution cell imaging. Biotechnol Prog 31, 1172 (2015).
- Welch, J. T. & Arden, N. S. Considering “clonality”: A regulatory perspective on the importance of the clonal derivation of mammalian cell banks in biopharmaceutical development. Biologicals 62, 16–21 (2019).
- Benchtop Microfluidic Cell Sorter: WOLF G2 | NanoCellect. https://nanocellect.com/products/wolf-g2-cell-sorter/?utm_ campaign=GS_Brand_BMM&utm_source=google&utm_medium=cpc&utm_content=text&utm_term=wolf%20 nanocellect&utm_campaign=GS_Brand_BMM&utm_source=adwords&utm_medium=ppc&hsa_acc=1686850884&hsa_net=adwords&hsa_grp=77679615039&hsa_ver=3&hsa_kw=wolf%20 nanocellect&hsa_tgt=kwd-819603803256&hsa_mt=e&hsa_ad=385078632457&hsa_src=g&hsa_cam=6542195443&gclid=Cj0KCQjwio6XBhCMARIsAC0u9aE1LpS4E3bgGVwZ6b422n1-eCCdJ7TfRhMciUHFhpuAIxWG6rW1wLEaApHvEALw_wcB
- Riba, J., Schoendube, J., Zimmermann, S., Koltay, P. & Zengerle, R. Single-cell dispensing and ‘real-time’ cell classification using convolutional neural networks for higher efficiency in single-cell cloning. Scientific Reports 2020 10:1 10, 1–9 (2020).
- Llufrio, E. M., Wang, L., Naser, F. J. & Patti, G. J. Sorting cells alters their redox state and cellular metabolome. Redox Biol 16, 381–387 (2018).
- Evans, K. et al. Assurance of monoclonality in one round of cloning through cell sorting for single cell deposition coupled with high resolution cell imaging. Biotechnol Prog 31, 1172–1178 (2015).
- Ng, W. L. et al. Controlling Droplet Impact Velocity and Droplet Volume: Key Factors to Achieving High Cell Viability in Sub-Nanoliter Droplet-based Bioprinting. Int J Bioprint 8, 1–17 (2022).
- The CellRaft AIR System enables workflows to image, sort and isolate viable single cells. https://cellmicrosystems.com/cellraft-air-systems/.
- Cell Microsystems - CytoSort Arrays. https://cellmicrosystems. com/cellraft-products/cytosort-arrays/.
- Cell Microsystem. CellRaft AIR System Brochure. (2021).
About Cell Microsystems
Making innovative tools that enable researchers to image, identify, and isolate viable single cells and clonal colonies
Researchers worldwide in the fields of CRISPR gene editing, oncology, stem cell biology, immunology and neurobiology use Cell Microsystems products, advancing sophisticated discovery across the life sciences.
The company’s CellRaft AIR® System addresses two widespread challenges facing scientists: the ability to actively select viable single cells or clonal colonies based on their phenotype, and match these cells to clonal expansion or molecular analyses. Cells are seeded, imaged, identified, and isolated on Cell Microsystem’s CellRaft® Arrays.
We have tested more than 100 Cell Lines with CellRaft Technology to demonstrate the high outgrowth efficiencies using our platform.
The company currently markets its products to researchers worldwide, and prides itself on being a customer-focused organization responsive to feedback and inspired to fuel deeper contributions to science.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.