The process of gene editing involves the transformation of a substantial number of cells, followed by the isolation of individual cells from the larger population to establish clonal colonies.
Due to the significant number of gene edits required in modern research and the labor-intensive workflow, there is a pressing need for automating the post-transformation cloning process.
This article presents data and protocols introducing the CellRaft AIR® System as a fully automated cloning platform. Through an imaging-based sorting approach, this system enables the monitoring of single cells for transformation-positive phenotypes and the maintenance of clonality as colonies develop.
The described protocols also provide evidence of enhanced cell viability, particularly with robust HEK293 cells, which exhibited nearly a threefold improvement in viability.
Previous studies have shown even more substantial viability enhancements for delicate cells, such as human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs). The system generates clonal colonies within 4-5 days, representing a substantial improvement over the months traditional workflows require.
Key highlights
- Automatically screen thousands of single cells
- Identify rare event-edited cells and track clonality
- Easily isolate monoclonal edited cell clones and organoids
- Get verifiable clones with more than 90% viability in days, not months
Generating clonal colonies
The CellRaft AIR® System and CellRaft Array offer an ideal solution for generating clonal colonies in transformation-based workflows, including gene editing. After transgenic or CRISPR/Cas9 elements are introduced, single cells are seeded onto the CellRaft Array.
Cells are periodically imaged to track the growth of a single cell within a designated microwell, eventually forming a clonal colony. Figure 1 illustrates an example of HEK293 cells transfected with a GFP-expressing plasmid.
Following transfection, single cells were seeded on the array, and the growth of clonal colonies was monitored over five days. Colonies of interest can be released from the CellRaft Array using the CellRaft AIR® System while still attached to the CellRaft.
The released colony is then deposited into a 96-well plate by the AIR System for continued expansion culture. Approximately 90% of the transferred colonies are typically viable after the transfer to the 96-well plate.
In the subsequent sections of this article, additional workflow details are explicitly provided for cells that have undergone CRISPR/Cas9 gene editing using an RNP-based workflow, where the Cas9 protein is pre-complexed with a guide RNA before introduction into the cell.
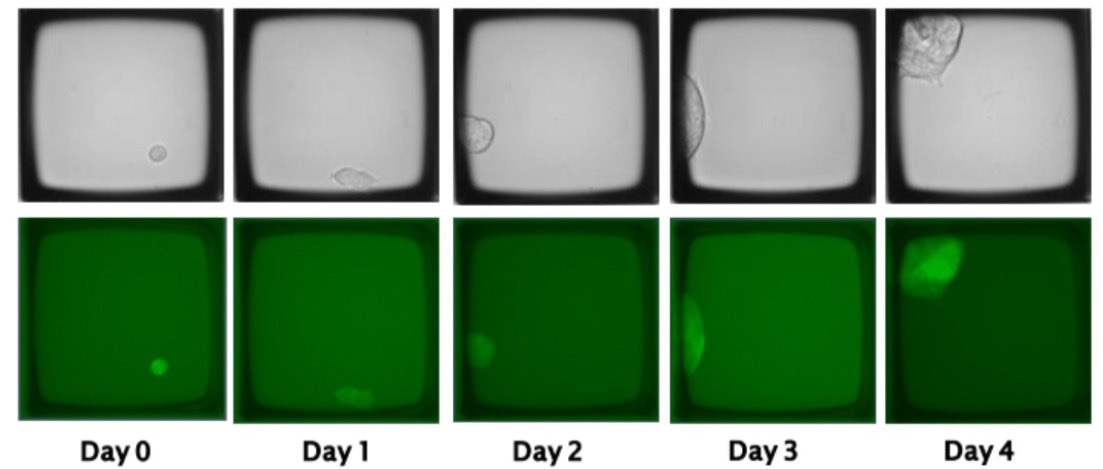
Figure 1. Cells transfected with a GFP-expressing plasmid can be seeded as single cells and tracked for the growth of clonal colonies on the CellRaft Array using the CellRaft AIR System. Image Credit: Cell Microsystems
The CellRaft AIR System
The CellRaft AIR System is a bench-top instrument with an internal microscope equipped with bright field and three-channel fluorescent imaging capabilities. This system utilizes the CellRaft® Array as a substrate for single-cell imaging and culture, along with the proprietary CellRaft Technology for isolating and collecting individual cells.
At its core, the technology consists of a disposable microwell array called the CellRaft Array. Cells are seeded and imaged on this array. To isolate single cells, a motorized needle penetrates the resealable elastomeric floor of the array, displacing the CellRaft from its respective microwell.
The CellRaft material is loaded with magnetic nanoparticles, enabling the retrieval of both the CellRaft and the attached cell using a magnetic wand.
The CellRaft AIR System leverages the same CellRaft Technology to automate fluorescent imaging, cytometric sorting, mechanical release, and collection operations. This automation significantly reduces hands-on time and greatly improves throughput.
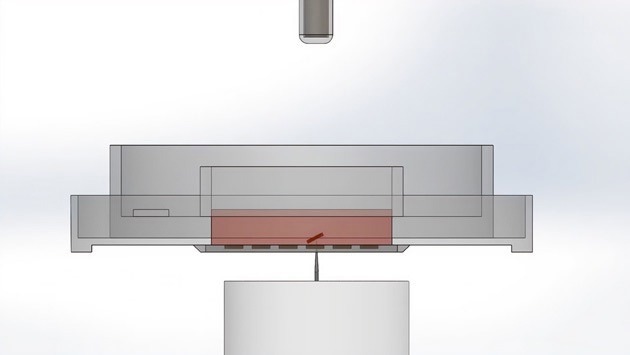
The desired microwell with cells is dislodged from the array. Image Credit: Cell Microsystems
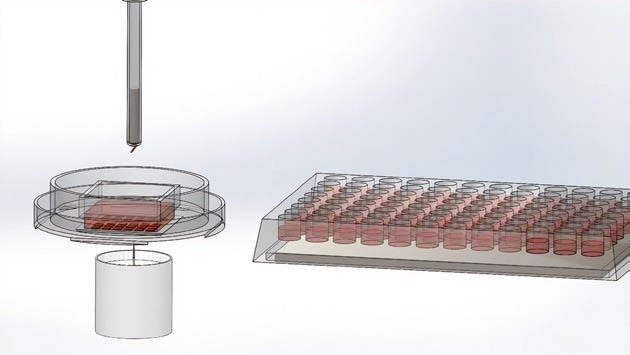
The wand picks up the microwell using a magnet. Image Credit: Cell Microsystems
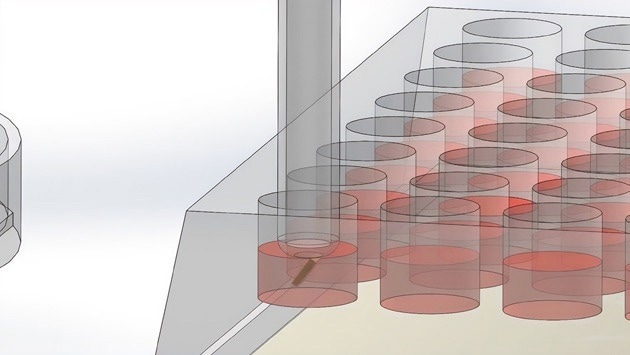
The wand places the microwell with the cells in the 96-well plate. Image Credit: Cell Microsystems
Comparing cloning workflows
To evaluate the suitability of the CellRaft AIR System for automated CRISPR cloning, a HEK293 cell line expressing both RFP and GFP was generated. Guide RNAs targeting the transgenic fluorescent proteins were then synthesized.
These guides were complexed with Cas9 using standard protocols provided by Integrated DNA Technologies (IDT) and introduced into cells using a Lonza Nucleofector. Four workflows were assessed for post-CRISPR cloning, as described below (with corresponding schematics in Figure 2).
The first workflow (FACS in Figure 2) followed a traditional flow sorting-based approach. Cells were seeded in 6-well plates and allowed to recover for 48 hours before undergoing sorting and isolation on a BD FACS Aria at the UNC-Chapel Hill Flow Cytometry Core facility.
In the second and third workflows, cells were also given a 48-hour recovery period in 6-well plates but then re-plated on CellRaft Arrays and isolated after 2 hours (Day 0 in Figure 2) or 24 hours of recovery (Day 1 in Figure 2), respectively.
In the final workflow (Direct-to-Array in Figure 2), cells were directly seeded onto the CellRaft Array, eliminating the 48-hour recovery period. In all cases, cells or colonies were isolated into 96-well plates, and viability was monitored.
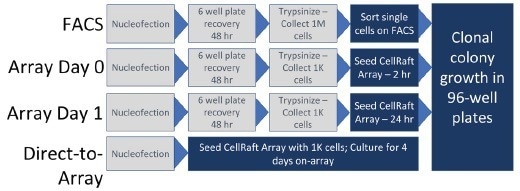
Figure 2. Schematic representation of the workflows evaluated for post-CRISPR cloning. Image Credit: Cell Microsystems
Monitoring clonality
Ensuring the growth of clonal colonies from gene-edited cells is a crucial aspect of the CRISPR workflow. The CellRaft AIR System allows for the imaging and tracking of individual cells throughout the colony formation process.
An example in Figure 3 showcases cells that underwent simultaneous knockout of GFP and RFP, resulting in the emergence of colonies from a single cell within four days. Moreover, the phenotypes of the knockout (GFP and RFP signal elimination) can be tracked over time, providing a unique capability for phenotypic CRISPR screening.
Control cells, which were transfected without a guide RNA, are also depicted, showing no alteration in fluorescent protein expression. Imaging enables the characterization of cellular phenotypes after gene editing, fulfilling the growing need for multiplex genome edits in large-scale screening studies.
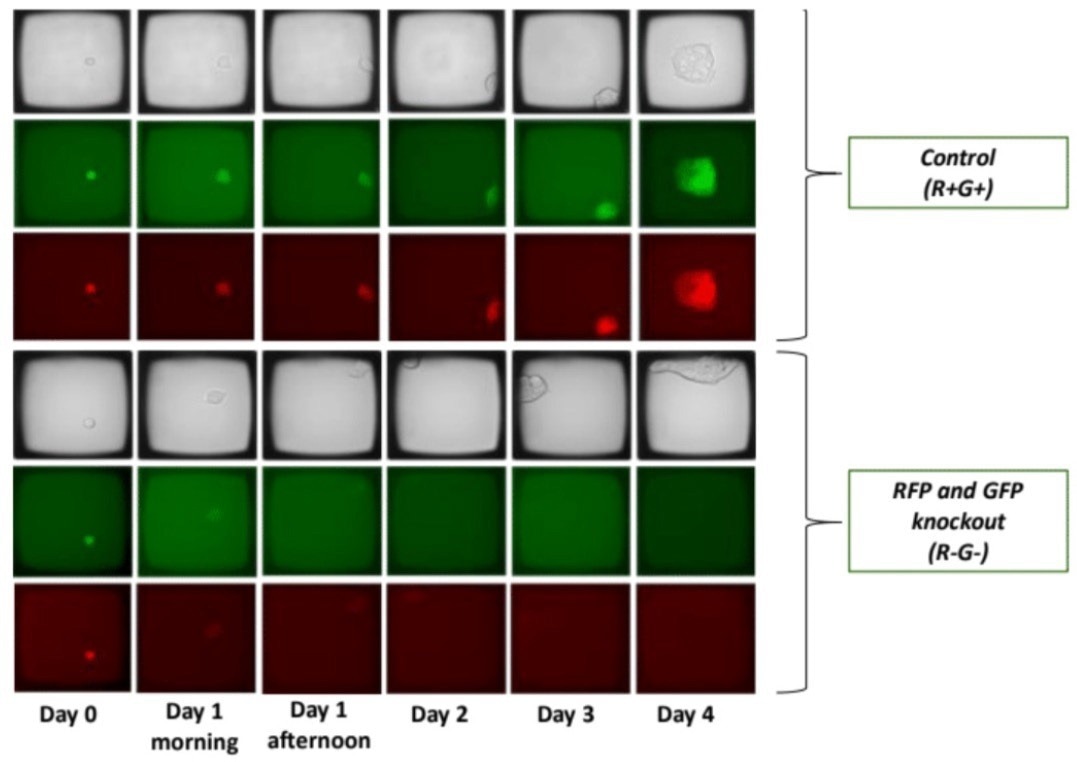
Figure 3. Single cell suspensions were seeded on the CellRaft Array and imaged on the CellRaft AIR System each day for 5 days (Day 0 = day of transfection). Each scan required 15 minutes, and arrays were placed back into a standard laboratory incubator between scans. By imaging, colonies were monitored for inclusion of extraneous cells to ensure clonality. Presence of cells in each microwell was confirmed with brightfield imaging. Image Credit: Cell Microsystems
Improved clonal viability
Cell sorting and single-cell culture often result in significant cell death. Even after isolating single cells in a culture dish, their viability tends to be low. This is typically attributed to the inability of a single cell to "condition" its culture environment through factors like secreted substances or exosome production.
On the CellRaft Array, single-cell viability is enhanced as the culture media volume is shared among thousands of cells across the array.
This feature offers an ideal solution for cloning experiments, especially with delicate cell types, such as primary cells, iPSCs, or hESCs, allowing cells to be recoverable as individuals while benefiting from the conditioning effects of thousands of cells in the culture environment.
After monitoring colonies for clonality using the CellRaft AIR System, the colonies were isolated following the protocols outlined in Figure 2. A 96-well plate containing either single cells (workflows 1-3) or clonal colonies (workflow 4) was prepared, resulting in 96 individual isolates for each workflow.
Subsequently, the viability of the colonies was monitored over a week. The viability data (Figure 4) indicates that the CellRaft AIR-based workflows improved viability by 2-3 times, with the highest viability observed in colonies recovered directly into 96-well plates after 48 hours on the CellRaft Array.
This increased viability is believed to result from omitting the multiple trypsin and re-plating steps associated with workflows 1-3.
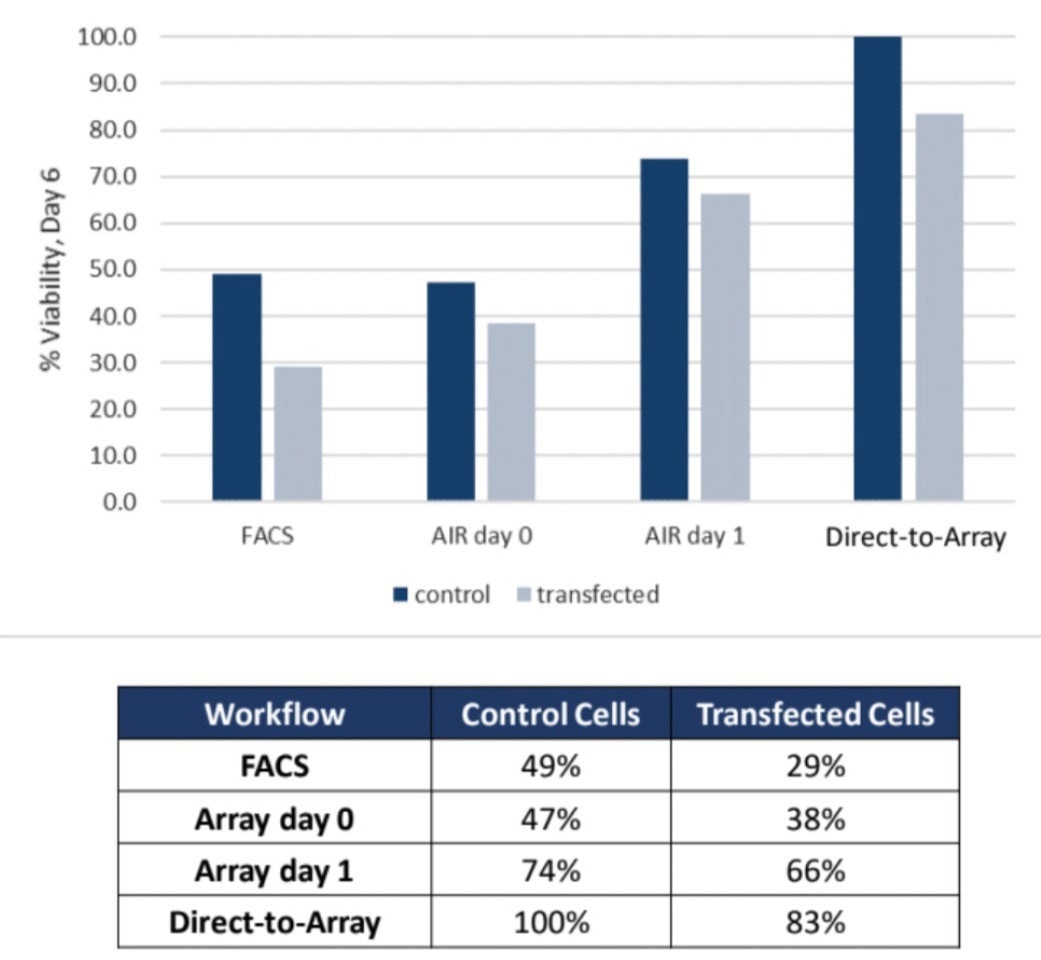
Figure 4. Clonal colonies collected from the CellRaft AIR System exhibit 2 to 3- fold greater viability compared to FACS-based workflows. Image Credit: Cell Microsystems
Efficiency of cloning
Generating stable clonal colonies with delicate cell types through genome editing can often take months. Service organizations typically require a turnaround time of 2-3 months, even for robust cells.
In contrast, the workflow presented here allows for the generation of stable clonal colonies within approximately 4-5 days. These colonies exhibit up to 90% viability and are verified for clonality through imaging-based tracking (Figure 5).
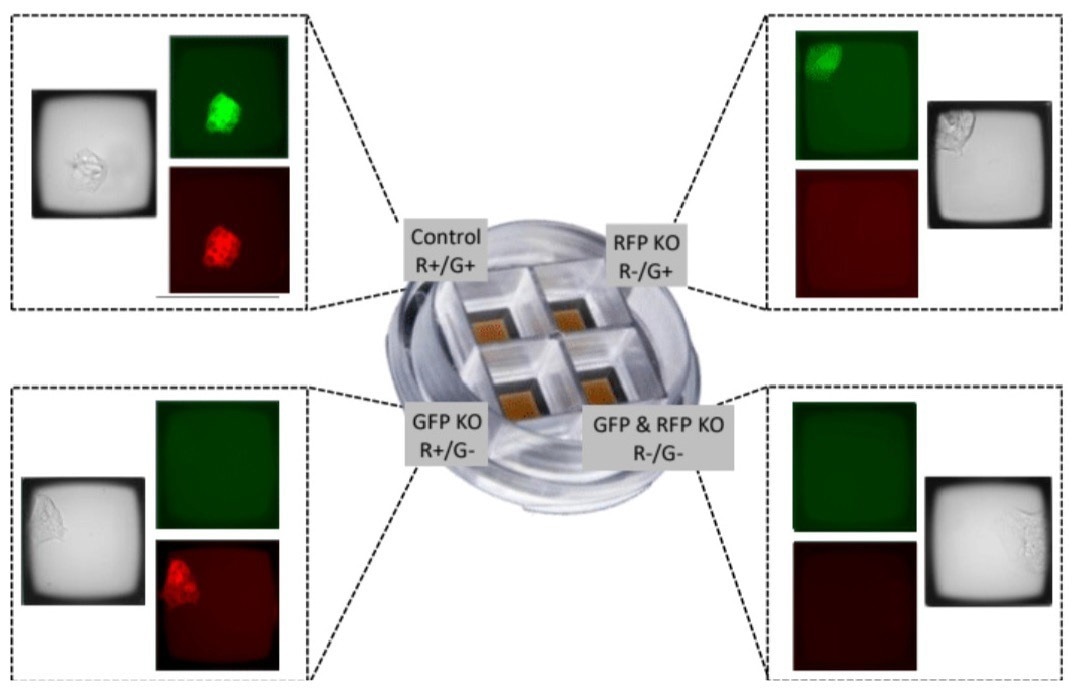
Figure 5. Use of the CellRaft QuadArray to isolate clonal colonies from four distinct CRISPR gene editing processes. Using the GFP/RFP-expressing model system, all four conditions were processed in parallel to generate both experimental and control gene editing cell lines in a single cell culture consumable. Given the 500 μL volume of each QuadArray well, reagent consumption was also reduced by 50-100 fold. Image Credit: Cell Microsystems
About Cell Microsystems
Making innovative tools that enable researchers to image, identify, and isolate viable single cells and clonal colonies
Researchers worldwide in the fields of CRISPR gene editing, oncology, stem cell biology, immunology and neurobiology use Cell Microsystems products, advancing sophisticated discovery across the life sciences.
The company’s CellRaft AIR® System addresses two widespread challenges facing scientists: the ability to actively select viable single cells or clonal colonies based on their phenotype, and match these cells to clonal expansion or molecular analyses. Cells are seeded, imaged, identified, and isolated on Cell Microsystem’s CellRaft® Arrays.
We have tested more than 100 Cell Lines with CellRaft Technology to demonstrate the high outgrowth efficiencies using our platform.
The company currently markets its products to researchers worldwide, and prides itself on being a customer-focused organization responsive to feedback and inspired to fuel deeper contributions to science.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.