This article discusses a collaborative case study using a new technology platform for fast and easy cell line development.
Monoclonal cell line development for NIH-3T3 cells expressing EBFP2 and HER-2
Developing innovative cell lines and disease models is a fundamental and essential tool for scientific exploration. Typically, this process involves the creation of a vector, such as a plasmid, virus, doggybone DNA, or transposon, to modify gene expression by either overexpressing or deleting genes.
The constructed vector is transfected or infected into cells of interest. Successful transgene integration is identified through flow cytometry, microscopy, or antibiotic selection (Figure 1).
Once a positive population has been restored, generating a suitable clone exhibiting the desired phenotype becomes essential to ensure uniformity in subsequent applications. Several methods can achieve a clonal population, including fluorescence-activated cell sorting (FACS), low-pressure dispensers, clone pickers, and limiting dilution.
However, utilizing these methods for developing cell lines often presents various challenges:
- Compromising cell viability, leading to inadequate colony growth from isolated clones
- Incurring high operational costs
- Requiring substantial amounts of consumables
- Being inefficient and labor-intensive for confirming clonality
- Demanding a high level of operator expertise and troubleshooting.
In this article, these issues are exemplified, emphasizing the significance of a novel technology/platform utilized to expedite scientific progress at Duke University's research laboratory in Durham, NC, USA.
The experiment aimed to generate an NIH3T3 cell line expressing human epidermal growth factor receptor 2 (HER2) and blue fluorescent protein (EBFP2) through two separate transductions.
According to the lead investigator, previous attempts to generate a double positive cell line using FACS failed to produce a pure polyclonal population, necessitating the creation of a viable and stable monoclonal line.
Leveraging the CellRaft AIR® System emerged as the preferred method for identifying clonal NIH-3T3 cells that were double positive for both transgenes.
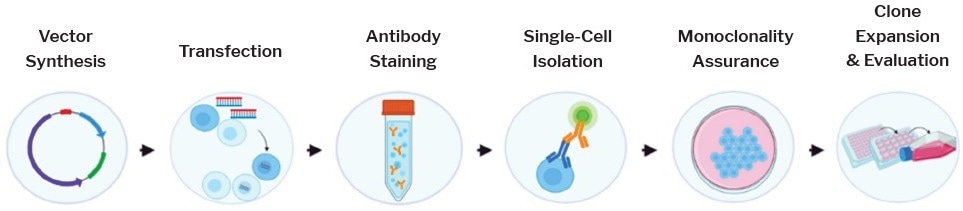
Figure 1. Cell Line Development Workflow. Traditional workflow involves synthesizing a vector with a gene of interest, transfecting cells with new genetic information, bulk staining a heterogenous population with an antibody of interest, isolating single cells with genes of interest, validating clonality, and expanding cells before transferring to cryovials for cryopreservation. Image Credit: Cell Microsystems
Key highlights
- Flow sorting failed to generate a stable double-positive cell population, even after multiple rounds of sorting.
- Rare double-positive clones could be identified using on-array antibody staining and analysis with the CellRaft AIR System’s software-guided imaging.
- The CellRaft AIR System successfully isolated 100% of the double-positive clones, with over 80% of the clones forming viable colonies for downstream applications
Scientific problem
A scientist modified NIH-3T3 fibroblast cells by introducing HER2 to establish a model representing HER2-driven breast cancer. To visualize the presence of HER2, the HER2-engineered cells were further transduced with EBFP2, a fluorescent marker.
The transduced cells were subjected to two rounds of FACS sorting to obtain a stable population exhibiting double positivity.
During routine cell culture passaging, it was observed that the expression levels of both EBFP2 and HER2 were drifting due to the presence of a heterogeneous population and the outcompeting nature of non-transduced cells.
To address these challenges, the researchers turned to Cell Microsystems and employed the CellRaft AIR System to overcome these limitations and obtain a suitable clone.
Experimental design
A heterogeneous bulk population of NIH-3T3 cells expressing EBFP2 and HER2 was seeded onto the CellRaft® Array. Over several days, the cells were imaged using the CellRaft AIR System, and parameters such as cell viability, morphology, fluorescence, and clonality were monitored.
Initially, software algorithms were used to select individual cells from the population. The blue fluorescence channel on the CellRaft AIR System facilitated the identification of EBFP2-positive cells.
On day six, live-staining with FITC-conjugated anti-HER2 was performed on the array to identify cells expressing HER2. By employing CellRaft CytometryTM, populations of single EBFP2+ cells on day 1 and EBFP2+/HER2+ cells on day 6 were created. The intersection of these two populations was used to identify monoclonal double-positive cells.
The double-positive clones were isolated into 96-well plates using the CellRaft AIR System, expanded, and subjected to off-array staining to verify HER2 expression. Finally, the validated clones were cryopreserved.
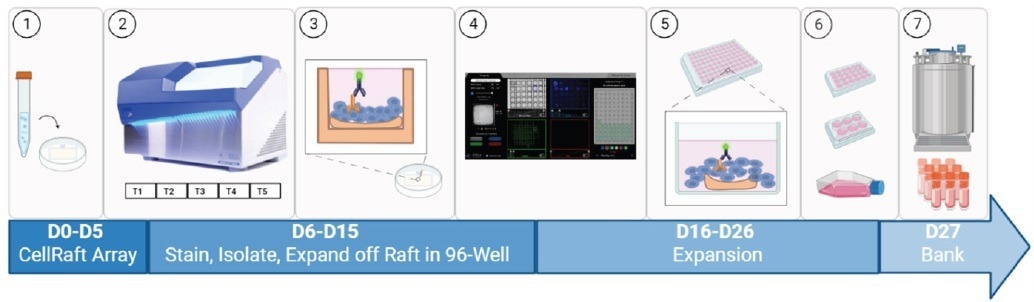
Figure 2. Cell Microsystems Workflow. (1) Heterogeneous cell mixture seeded onto a 200uM Single CellRaft Array. (2) CellRaft Array scanned on CellRaft AIR System over several timepoints. (3) CellRaft Array is live stained with Anti-HER2. (4) CellRaft Cytometry identifies rafts of interest containing the population of cells double positive for selected markers of interest. (5) CellRafts are isolated into 96-well collection plates and expanded into a sizable colony. (6) Monoclonal colonies are expanded and transferred into T-flasks. (7) Expanded clones are transferred to cryovials for cryopreservation as a master cell bank. Image Credit: Cell Microsystems
Results
Following an initial FACS sorting, around 10% of the transduced NIH-3T3 cells exhibited double positivity for EBFP2 and HER2 (Figure 3A). The population of 10% double-positive cells was expanded and re-sorted, resulting in an overall 89% population expressing both EBFP2 and HER2 (Figure 3B).
Despite the reported 89% purity achieved through FACS analysis, it was observed that the expression of the two transgenes was not stable after thawing and passaging, with HER2 expression varying throughout the passage.
To determine the percentage of cells exhibiting double positivity, the bulk population was stained with FITC-conjugated AntiHER2. While most cells visibly showed EBFP2 expression (Figure 4B), the actual population positive for HER2 was less than 10% (Figure 4C).
For the acquisition of a monoclonal clone that expressed both EBFP2 and HER2, the heterogeneous population was seeded on a CellRaft Array. After cell attachment, the arrays were imaged over six days using the CellRaft AIR System (Figure 5-6).
Using CellRaft Cytometry, it was convenient to identify CellRafts that harbored the desired clones of interest. Figure 5 illustrates the presence of CellRafts that contained a solitary EBFP2+ cell at time point 1, evolving into a uniform EBFP2+/HER2+ colony by time point 6, representing the rare occurrence of double positives.
In contrast, polyclonal CellRafts could be easily distinguished and disregarded, ensuring both gene expression and clonality through image-based phenotyping (Figure 6). Out of the 6,000 cells screened on a single CellRaft Array, a mere 76 double-positive clones were found, while over 400 clones were identified as EBFP2+/HER2- (Figure 7).
The researchers isolated 33 CellRafts, which contained the rare double-positive clones, with an isolation efficiency of 100% and an outgrowth efficiency of over 80%.
This underscores the capability of the CellRaft AIR system to effectively identify and isolate these rare double-positive clones, which comprised less than 1.5% of the entire population. Upon isolation and expansion, all the clones were subjected to re-testing, confirming their double positivity for both EBFP2 and HER2 (Figure 8).
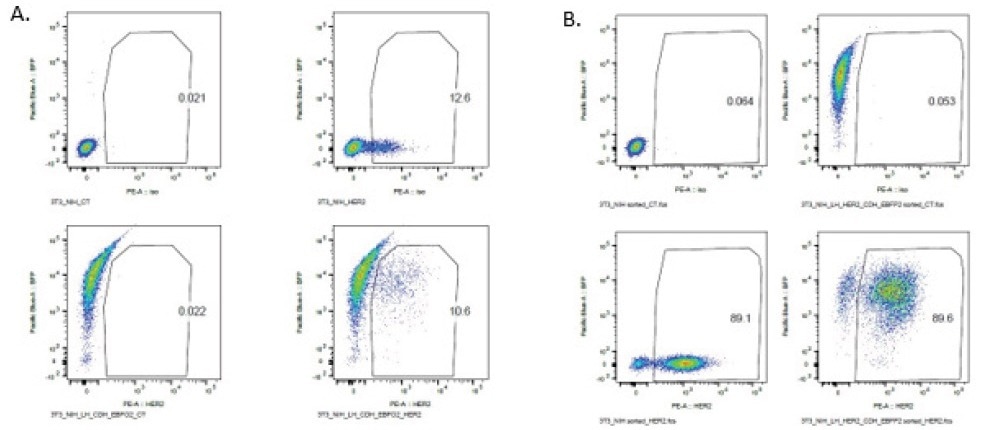
Figure 3. FACS analysis of NIH-3T3 HER2 EBFP2 cells NIH-3T3 cells were stained for HER2 expression and sorted for EBFP2 and HER2 double positivity. After an initial bulk sort (A), 10% of the population was double positive, and a re-sort (B) of that pre-sorted 10% yielded a population that was 89% double positive. Image Credit: Cell Microsystems
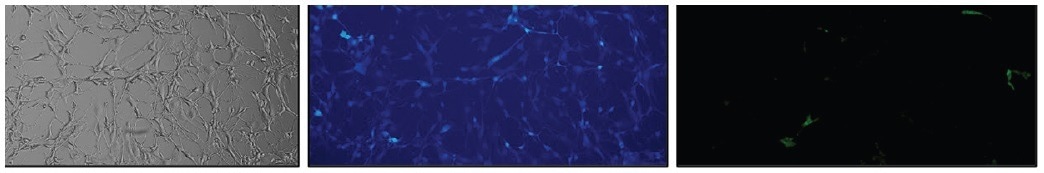
Figure 4. NIH/3T3 EBFP2+ HER2+ cells stained with Anti-HER2-FITC. (A) Brightfield, (B) EBFP2 expression (blue), and (C) HER2 expression (green). 10X Magnification. Image Credit: Cell Microsystems
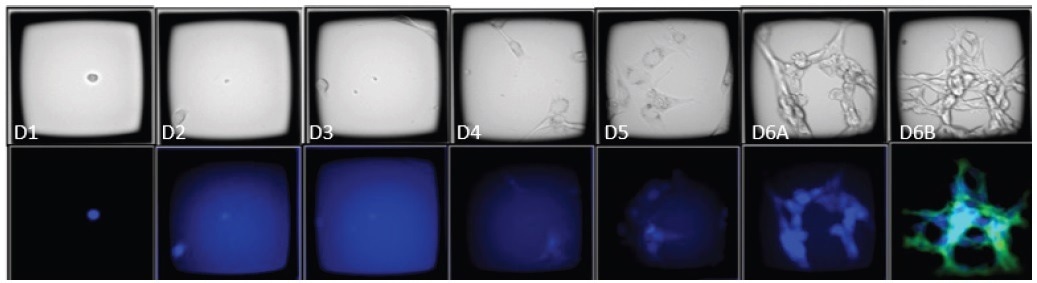
Figure 5. Time course images of a CellRaft containing an EBFP2+/HER2+ clone. The CellRaft arrays were imaged in brightfield and fluorescence 4 hours post-seeding and every 24 hours until colony formation. On day 6, the array was stained with Anti-HER2-FITC to visualize HER2 expression (green). 10X Magnification. Image Credit: Cell Microsystems
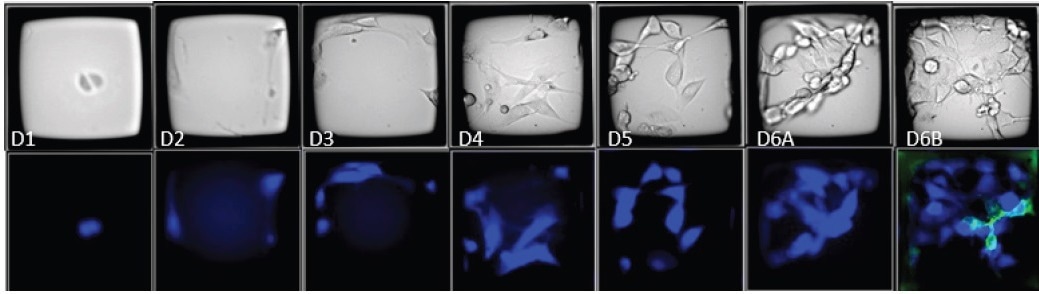
Figure 6. Time course images of a CellRaft containing polyclonal EBFP2+/HER2- and EBFP2+/HER2+ cells. The CellRaft arrays were imaged in brightfield and fluorescence 4 hours post-seeding and every 24 hours until colony formation. On day 6, the array was stained with Anti-HER2-FITC to visualize HER2 expression (green). 10X Magnification. Image Credit: Cell Microsystems
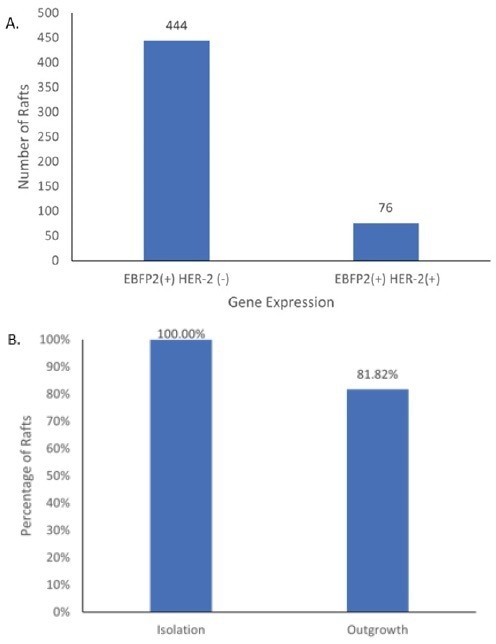
Figure 7. Identification and Isolation of NIH-3T3 EBFP2+/HER+ clones (A) Using CellRaft Cytometry, CellRafts containing single-cell derived colonies that were EBFP2+ and HER2+ were identified (B) CellRafts containing EBFP2+/HER2+ clones were isolated using the CellRaft AIR System and isolation and outgrowth efficiencies were determined by manual observation. Image Credit: Cell Microsystems
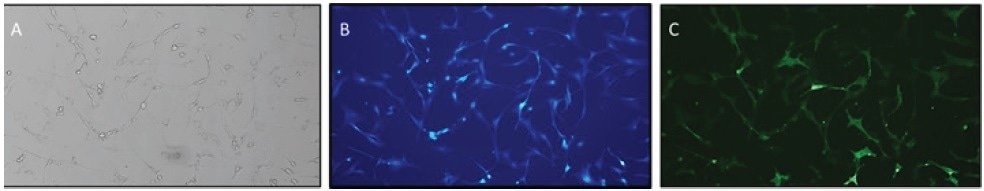
Figure 8. NIH-3T3 EBFP2+/HER2+ clone expanded off the CellRaft. After isolation and expansion, the double positive clones were stained with Anti-HER2-FITC to confirm gene expression. (A) Brightfield, (B) EBFP2 expression (blue), and (C) HER2 expression (green). 10X Magnification. Image Credit: Cell Microsystems
Discussion
An ongoing challenge in cell line development involves establishing stable cell lines that maintain consistent gene expression throughout thawing, passaging, and experimental procedures. This challenge becomes even more pronounced when multiple transgenes are simultaneously transduced, and the expression of each gene is not correlated.
Traditional approaches like FACS often yield heterogeneous cell populations with varying levels of the genes of interest, alongside wild-type cells, potentially obscuring or diluting downstream analysis.
Moreover, this heterogeneity introduces a survival competition within the population, leading to drift and inconsistent results, as evident from the aforementioned case study data.
The initial NIH-3T3 EBFP2+/HER2+ cell line achieved only 10% double positivity after transduction, and subsequent rounds of FACS cleanup yielded a polyclonal line with only 89% purity.
However, immunofluorescence characterization revealed that approximately 1.5% of the expanded population exhibited double positivity.
The identification, recovery, and single-cell expansion of this small percentage using FACS or limiting dilution would prove immensely challenging, requiring the screening of millions of cells with a low success rate.
Conclusion
Given the urgent requirement to swiftly develop genetically edited analytical cell lines for the advancement of drug and disease research, the research community seeks solutions to overcome obstacles hindering the simple and effective creation of clonal cell lines.
By using the CellRaft AIR System, the team achieved the screening of numerous cells within a single consumable, swiftly identifying the scarce double-positive clones in a matter of weeks rather than months.
In total, the process from cell seeding to clonal outgrowth took a mere 16 days, and viable cell banks of the verified clones were cryopreserved within 27 days.
Hence, the employment of CellRaft AIR® Technology facilitates the efficient and expedited development of monoclonal cell lines, even for cells that are rare or pose challenges, surpassing the effectiveness of traditional methods.
The generation of these double positive clonal lines saved my lab of a lot of time and effort, allowing the rapid generation of multiple highly stable clones that had persistent expression for in vitro and in vivo assays. This proved superior to our flow-based methodologies, which were never as transgene positive or stable (in terms of expression) over multiple passages.”
Zachary Hartman, Ph.D. Director, Center for Applied Therapeutics. Associate Professor, Departments of Surgery, Pathology, and Immunology, Tumor Immunology and Immunotherapeutics Laboratory. Duke University. Review of the CellRaft Air System.
References and further reading
- Biorender. BioRender. (n.d.). Retrieved October 7, 2022, from https://biorender.com/
- Molecular Devices. (2019, September 20). Optimize your high-value cell lines. San Jose.
About Cell Microsystems
Making innovative tools that enable researchers to image, identify, and isolate viable single cells and clonal colonies
Researchers worldwide in the fields of CRISPR gene editing, oncology, stem cell biology, immunology and neurobiology use Cell Microsystems products, advancing sophisticated discovery across the life sciences.
The company’s CellRaft AIR® System addresses two widespread challenges facing scientists: the ability to actively select viable single cells or clonal colonies based on their phenotype, and match these cells to clonal expansion or molecular analyses. Cells are seeded, imaged, identified, and isolated on Cell Microsystem’s CellRaft® Arrays.
We have tested more than 100 Cell Lines with CellRaft Technology to demonstrate the high outgrowth efficiencies using our platform.
The company currently markets its products to researchers worldwide, and prides itself on being a customer-focused organization responsive to feedback and inspired to fuel deeper contributions to science.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.