With the current COVID-19 pandemic having already claimed hundreds of thousands of lives worldwide, scientists are working to bring out effective antiviral strategies that can be quickly deployed, not only for the current scenario but to counter future threats.
The Principle
The current study is based on the principle of designing a protein surface that is identical to that cell surface targeted by the SARS-CoV-2 virus but with a higher binding affinity for the viral RBD than the host cell. This will also need to inhibit viral attachment to the cell. The end result will be that these “de novo designer proteins can outcompete the viral interaction and act as neutralizing agents.” Moreover, the very design of the decoy ensures that mutational escape is impossible since any mutation that weakens viral binding to the decoy will automatically also weaken its ability to bind to the receptor surface of which the decoy is a copy.
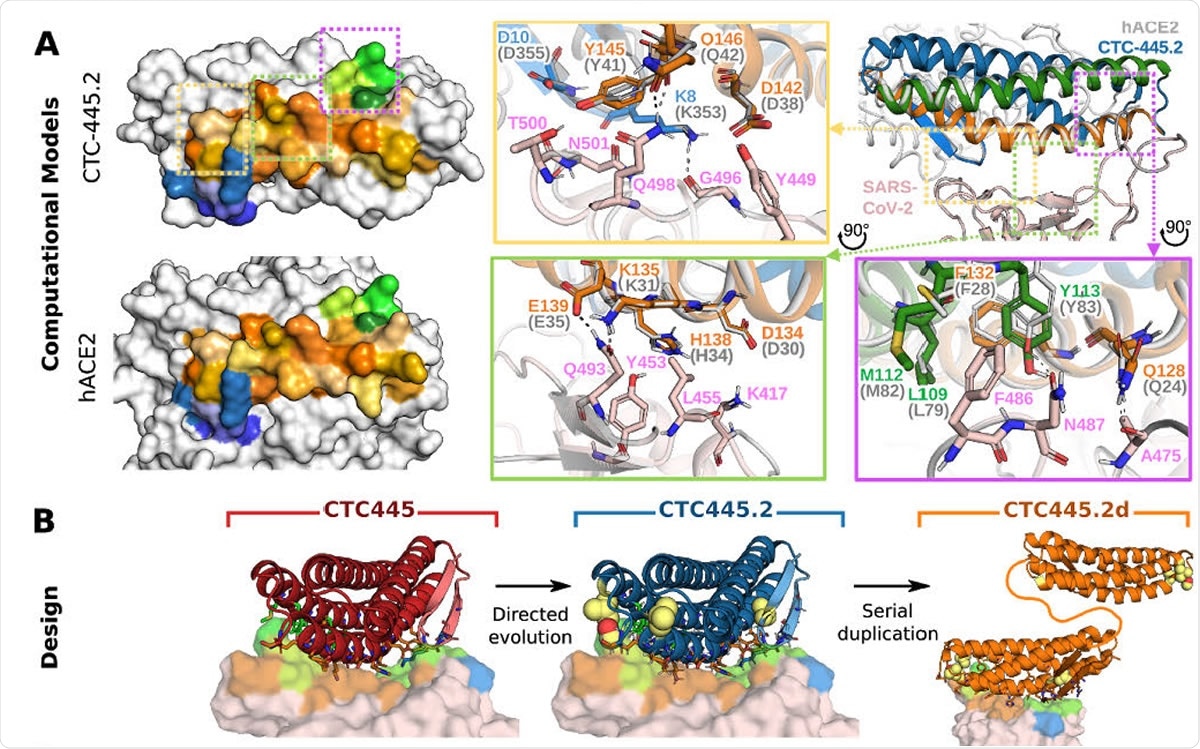
Binding, stability and structure of the de novo protein decoys CTC-445, CTC-445.2 and CTC-445.2d. A) Computational models of CTC-445.2. Left, comparison of the SARS-CoV-2 binding interface surface of CTC-445.2 (top) and hACE2 (bottom). CTC-445.2 is designed to present the same binding interface as the one that SARS-CoV-2 targets in hACE2, down to the level of individual interactions. Residues from the binding motifs: H1 are shown in shades of orange, residues from H2 in shades of green and residues from EE3 in shades of blue. The boxes show detailed structural comparison of the interfaces between CTC-445.2 and hACE2 with SARS-CoV-2 RBD. The relaxed complex of hACE2 with SARS-CoV-2 RBD (dark and light gray, respectively; PDB: 6M17) are aligned to the model of the relaxed compex of CTC-445.2 and SARS-CoV-2 RBD (pink). Hydrogen bond interactions are indicated by black dashed lines; B) Design models of CTC-445, CTC-445.2 and CTC-445.2d. CTC-445.2 contains 5 mutations that were guided by directed evolution experiments. CTC-445.2d is a bivalent variant composed of two CTC-445.2 subunits linked by a 17-mer flexible GS linker (sequence -GGGGSGGSGSGGSGGGS-);

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Process
The scientists first identified the characteristic repeating structures that make up the protein surface actually bound to by the viral RBD. This is based on three structures made publicly available, reflecting the possible structure of the RBD-ACE2 complex for SARS-CoV-1 and SARS-CoV-2. They picked out the four structural components that make up the binding interface of the ACE2 protein and built the de novo decoy with three of them – two long alpha-helices and one short beta-hairpin. They avoided any part of the biologically active part of the molecule, such as the enzymatic catalytic site or the site of interaction with the cell membrane.
The target elements were then mounted on a new standalone support structure that allows proper folding of the binding site into the globular shape required and stabilizes the interface. The Rosetta software was used to develop about 35,000 fully connected protein topologies with all these elements, without altering even the conformation of the amino acids of the target binding interface. The amino acid sequences were then generated such that they could fold into the target structures. The designs were then evaluated using an automatic filter, and the top 196 were tested for binding to the virus. They found that one molecule, which they named CTC-445.2d (ConquerTheCorona445.2duo), had a nanomolar and very specific affinity of binding to the SARS-CoV-2 RBD, but failed to inhibit its binding to ACE2.
The researchers then carried out targeted mutagenesis to incorporate a planned combination of the most useful mutations found to stabilize the protein and further enhance its affinity of binding. This resulted in CTC-445.2, with five substituted amino acids, none at the binding interface. This was found to be very stable and a potent inhibitor of infection in vitro. Finally, they performed a domain duplication to make it bivalent. This step resulted in CTC-445.2d, with tenfold binding affinity and 100 times the neutralizing capacity of the earlier version of the protein.
The Protein
CTC-445.2d is a 160 amino acid protein which binds the SARS-CoV-2 RBD at nanomolar concentrations, is very stable, and is also reactive for SARS-CoV-1 RBD. Though it is such a strong competitor of viral infection, cell viability remains unaffected as does the enzymatic activity of ACE2. Complete neutralization of viral infection was shown in three in vitro systems, including in a lung epithelial cell-derived system Calu-3.
Further, its efficacy was greatest when present throughout the course of infection, that is, when it was incubated along with the virus and in cell media, indicating that it inhibited the infection by extracellular neutralization. The researchers also instilled a high dose of the protein intranasally to an experimental mouse model, and found that the molecule could be found in a fully functional form for over 24 hours, and also detectable in blood. This points to the possibility of systemic exposure as well at some level.
The Advantages
The use of soluble decoy proteins is quite different from that of vaccines, small molecule inhibitors or neutralizing antibodies, and overcomes the problem of mutational escape. These molecules also avoid the stability and unwanted biological effects, as well as potential autoimmunity, implicit in the use of natural proteins. Being quite different from natural proteins in their amino acid sequence and structure, they are unlikely to elicit autoimmune responses.
These small molecules are easy to produce at large scale in available bacterial systems, are stable over a wide range of conditions, and can be further refined to increase their potential for affinity and neutralization potency. The decoys are functionally resilient to viral escape mutations as well. The underlying principle is validated by the cross-reactivity of the decoy proteins with SARS-CoV-1, because the differences in the RBD of the two viruses involve the binding interface at least at 17 sites but fail to prevent neutralization of both by the same protein.
The researchers say, “Using our novel platform, in less than ten weeks, we engineered, validated, and optimized de novo protein decoys of human angiotensin-converting enzyme 2 (hACE2).” These very stable, high-affinity small proteins bind to the viral RBD and prevent it from attaching to the host cell. They predict that by further developing this novel, accurate and rapid technique, ït should be possible to rapidly develop “other therapeutic de novo protein decoys, not limited to neutralizing viruses, but to combat any agent that explicitly interacts with cell surface proteins to cause disease.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources