Protein-protein interactions form the basis of cell structure and function, and the activity of a protein is often regulated through interaction with other proteins. Consequently, protein interactions have been thoroughly studied in order to further understanding of how the different systems of the body function and communicate with each other.
In addition, they often provide important information in disease states when a system is not working as it should. Furthermore, the modulation of protein-protein interactions using chemical or biological agents has become an important strategy in the development of novel therapeutics.
The precision required of protein-protein interactions in order for them to achieve the desired structure or function suggests that an evolutionary process must have occurred to develop protein-protein interactions that are optimized for their particular purpose.
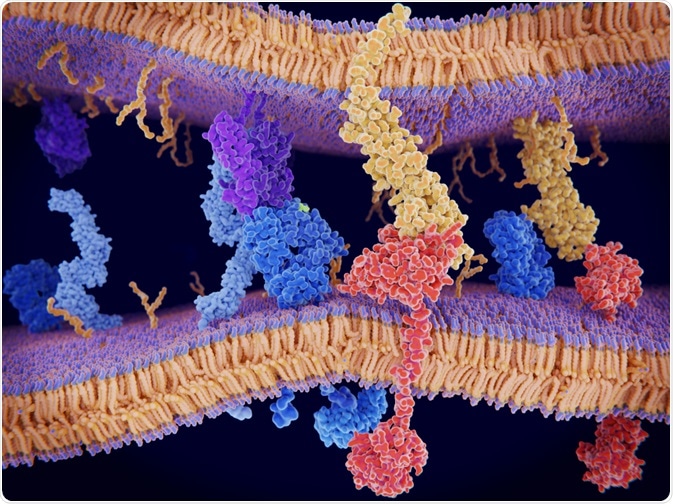
Image Credit: ©Juan Gaertner/Shutterstock.com
Protein-protein interactions
Protein-protein interactions are the high-specificity contacts between two or more protein molecules. Such interactions control virtually all vital cellular processes, including gene expression, proliferation, intracellular communication, and apoptosis. They are determined by the sequences of the proteins involved and are achieved through a range of electrostatic forces1.
There is a complex web of protein-protein interactions that often involve competing interactions, and all are affected by spatial and temporal cues to ensure the desired regulation of the proteins.
Evolution of protein-protein interactions
The nature of the protein-protein interactions seen today is likely to have evolved over tens or hundreds of millions of years to achieve optimum regulatory control. As a new protein-protein interaction arose as a result of, say a point mutation or gene duplication, it would lead to a different resultant effect on the biochemistry of the cell than was intended.
Usually, the altered effect would not have been as beneficial to the cell as the intended effect, and so did not persist. However, on some occasions, the new interaction will have proved to have beneficial consequences on cell functioning and will have been retained in the population by natural selection.
Despite the critical importance of protein-protein interactions, surprisingly little is known about the molecular details of the evolution of new protein-protein interactions in nature. With recent advances in genomic manipulation techniques, ancient proteins from evolutionary ancestors are being reconstructed from the sequences of species existing today. In this way, it possible to retrospectively study possible evolutionary pathways that led to the protein-protein interactions we know today2.
Most recently, changes in the structure and dynamics during the evolution of a protein-protein interaction were investigated for the intrinsically disordered CREBBP (CREB-binding protein) interaction domain (CID) and nuclear coactivator binding domain (NCBD) from the transcriptional coregulators NCOA (nuclear receptor coactivator) and CREBBP/p300, respectively3.
The CREBBP/p300 and NCOA proteins contain histone acetyltransferase domains and are involved in transcriptional regulation, for example, by activating steroid receptors4. The affinity of the CID/NCBD protein-protein interaction seen in organisms today is the same as that seen in an early ancestor of present-day fish living around 500 million years ago. However, the affinity was much lower and associated with greater conformational heterogeneity in species present around 600 million years ago.
The tighter association achieved by a new protein-protein interaction between NCOA and CREBBP/p300 probably resulted in a more efficient transcription, which was sufficiently beneficial to be retained by natural selection in an ancestral population leading to present-day deuterostome animals5.
To explore the nature of this evolutionary change, the structures of three historical CID/NCBD complexes were obtained by nuclear magnetic resonance (NMR) using Bruker spectrometers (600, 700, and 900 MHz, 1H frequencies), equipped with triple-resonance cryogenic temperature probes at 298 K. The dynamics of the protein-protein interaction were then studied using relaxation dispersion NMR spectroscopy3.
The findings revealed that the most ancient Cambrian-like CID/NCBD complex lacked some of the secondary structure apparent in the more recent forms of the complex. For example, one helix was missing altogether.
In addition, there has been a reduction and reshuffling of the backbone dynamics from the low-affinity ancestral complex to the younger Ordovician-Silurian/extant human complexes. These motions were reduced in the Ordovician-Silurian CID/NCBD complex and further redistributed in the extant human CID/NCBD complex.
Isothermal calorimetry experiments showed that complex formation is enthalpically favorable and that affinity is modulated by a largely unfavorable entropic contribution to binding.
These data demonstrate how changes in structure and motion conspire to shape affinity during the evolution of a protein-protein complex and provide direct evidence for the role of structural, dynamic, and frustrational plasticity in the evolution of interactions between intrinsically disordered proteins. Such rearrangements cannot easily be predicted, but the advanced imaging techniques available today have enabled snapshots of evolutionary change to be captured,
References
- Berggård T, et al. Proteomics. 2007;7(16):2833‑2842.
- Harms MJ and Thornton JW. Nat. Rev. Genet. 2013;14:559–571.
- Jemth P, et al. Structure and dynamics conspire in the evolution of affinity between intrinsically disordered proteins. Science Advances 2018;24(Vol. 4):no. 10, eaau4130. http://advances.sciencemag.org/content/4/10/eaau4130
- Leo C and Chen JD. Gene 2000;245:1–11.
- Hultqvist G, et al. eLife 2017;6:e16059.
About Bruker BioSpin Group
The Bruker BioSpin Group designs, manufactures, and distributes advanced scientific instruments based on magnetic resonance and preclinical imaging technologies. These include our industry-leading NMR and EPR spectrometers, as well as imaging systems utilizing MRI, PET, SPECT, CT, Optical and MPI modalities. The Group also offers integrated software solutions and automation tools to support digital transformation across research and quality control environments.
Bruker BioSpin’s customers in academic, government, industrial, and pharmaceutical sectors rely on these technologies to gain detailed insights into molecular structure, dynamics, and interactions. Our solutions play a key role in structural biology, drug discovery, disease research, metabolomics, and advanced materials analysis. Recent investments in lab automation, optical imaging, and contract research services further strengthen our ability to support evolving customer needs and enable scientific innovation.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.