Glioblastoma mutliforme (GBM) represents the most dangerous and malignant primary brain tumor.
A combination of resident microglia and peripheral infiltrating macrophages accounts for up to 50% of non-neoplastic cells present in a GBM, with these tumor-associated macrophages playing a role in proliferation, angiogenesis and immunosuppression.
They can also impact the efficacy of chemo-, radio- and immunotherapies when they are used to treat GBM.
Research has shown that some cancer-specific interactions have been linked to either microglia or macrophages, meaning it is necessary to develop a means of delineating the two populations.
The use of multiplex immunofluorescence offers a potential solution to this task, with the technical benefit of facilitating the profound phenotyping and analysis of the spatial organization of cells in the tumor microenvironment.
This article looks at ongoing work to design a multiplex immunofluorescence protocol able to differentiate between microglia and infiltrating M1/M2 macrophages in GBM and suitable for use in situ.
Methods
- Panel design and validation were done using a HISTOPROFILE® Neuro M1/M2 panel.
- Targets identified were:
- GFAP/TMEM119/CD68/c-Maf/CD163
- Microglia: TMEM119+
- M2: CD68+/CD163+/c-Maf+/TMEM119-
- M1: CD68+/CD163-/c-Maf-/TMEM119-
- Tumor: GFAP+
- Human Glioblastoma FFPE Blocks were acquired from the Cerba Research biobank in Montpellier.
- A sequential multiplex protocol with Opal® (Akoya Biosciences) fluorophores was developed on the BOND RX® (Leica) slide stainer.
- It was then possible to acquire whole slide images with the VECTRA® Polaris™ (Akoya Biosciences) slide scanner.
- Image Analysis was performed using the HALO® (Indica Labs) Highplex module
- The total numbers of Microglia, M2 and M1 macrophages were quantified.
- Infiltration was investigated at the tumor/non-tumor interface.
HISTOPROFILE® Neuro M1/M2: Phenotyping of three independent samples
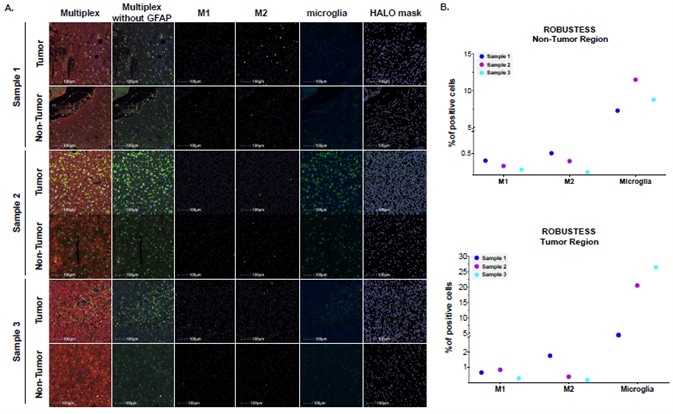
Image Credit: Cerba Research
Results
HISTOPROFILE®-Neuro M1/M2 were collected on three independent GBM samples.
Figure A displays example images of the tumor and non-tumor regions of the three samples. These have been stained with the HISTOPROFILE® Neuro M1/M2 panel: GFAP (red), TMEM119 (green), CD68 (yellow), CD163 (white), c-Maf (orange).
Representative images from left to right feature: merged multiplex, macrophage phenotyping targets (TMEM119, CD68, CD163, c-Maf), M1 HALO® masks, M2 HALO® masks, microglia (TMEM119), and microglia HALO® masks.
Figure B features graphs displaying the percentage of positive cells for M1 (CD68+/CD163-/c-Maf-/TMEM119-), M2 (CD68+/CD163+/c-Maf+/TMEM119-) and microglia (TMEM119+) in the three samples.
It is possible to observe notable differences in these three samples between non-tumor (upper graph) and tumor (lower graph) regions.
HISTOPROFILE® Neuro M1/M2: Panel validation
Using a simplex protocol, simplex slides were stained for individual biomarkers. These were then compared against a serial section that had been stained with the multiplex HISTOPROFILE® Neuro M1/M2 IHC panel.
It was then possible to perform analysis with HALO® to determine staining concordance between the multiplex slide and simplex slides.
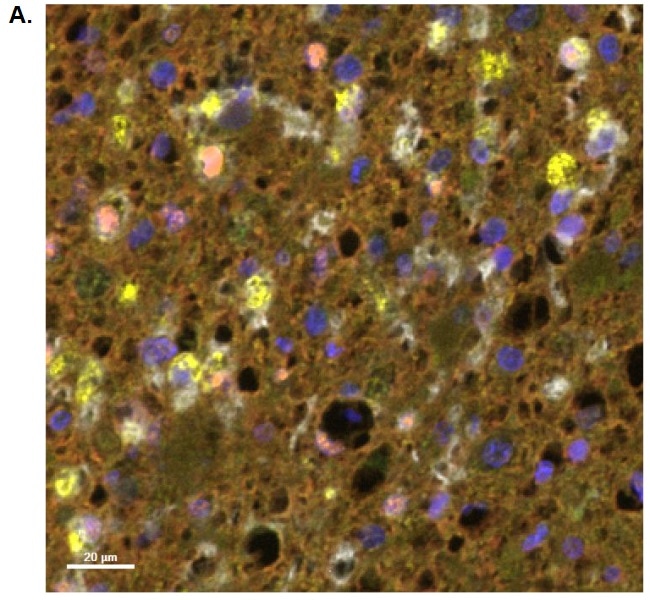
Image Credit: Cerba Research
Figure A displays an example image of a HISTOPROFILE® Neuro M1/M2 multiplex stained slide: GFAP (red), TMEM119 (green), CD68 (yellow), CD163 (white), c-Maf (orange).
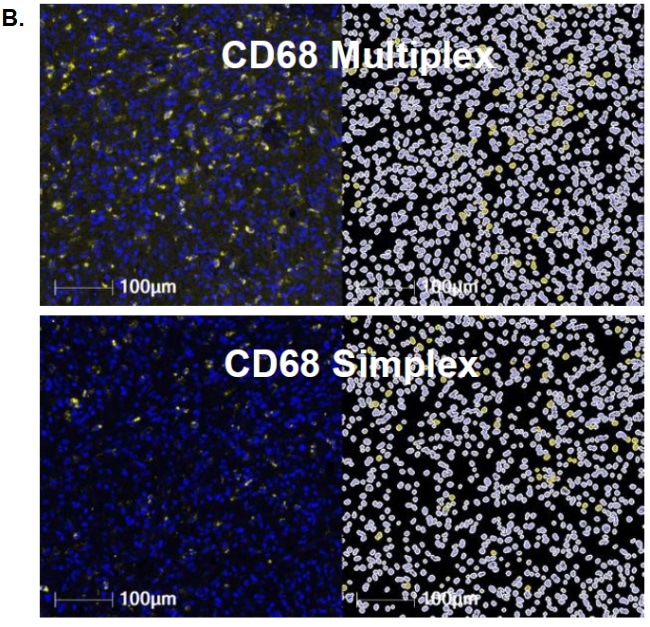
Image Credit: Cerba Research
Figure B features example images that correspond to the multiplex slide for CD68 (top) and the simplex slide (bottom). It also shows the corresponding HALO® masks, which confirm the identification of positive cells.
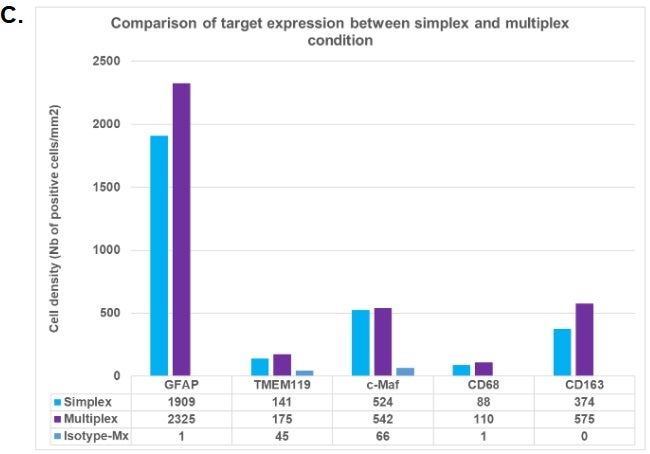
Image Credit: Cerba Research
Figure C provides results from the simplex versus multiplex panel analysis. A comparison was made between the percentage of positive cells from the simplex slide (blue bars) and the multiplex slide (purple bars).
This confirmed that satisfactory results were acquired for the HISTOPROFILE® Neuro M1/M2 panel (simplex/multiplex ratio: 1.3 to 0.7). Using intra-run repeatability analysis and inter-run reproducibility analysis, it was possible to determine the protocol’s precision.
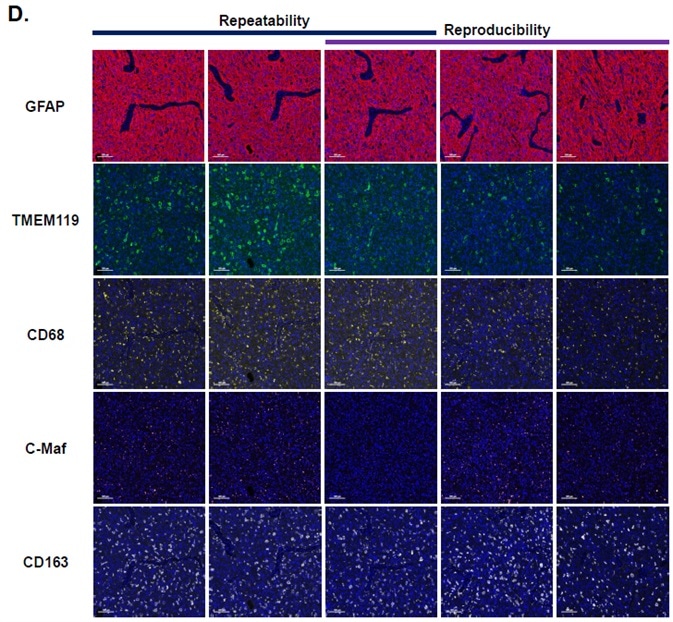
Image Credit: Cerba Research
Figure D features example images from the reproducibility and repeatability slides.
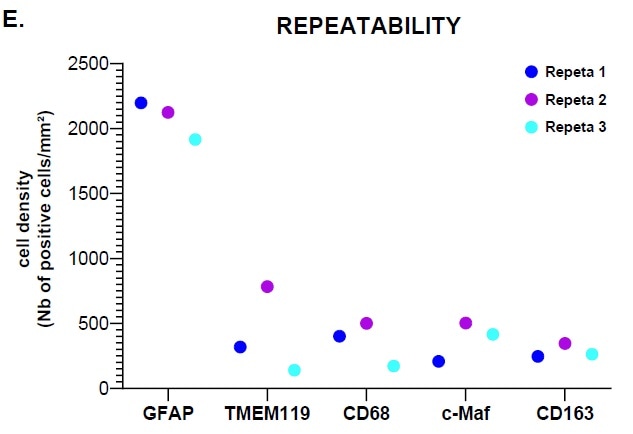
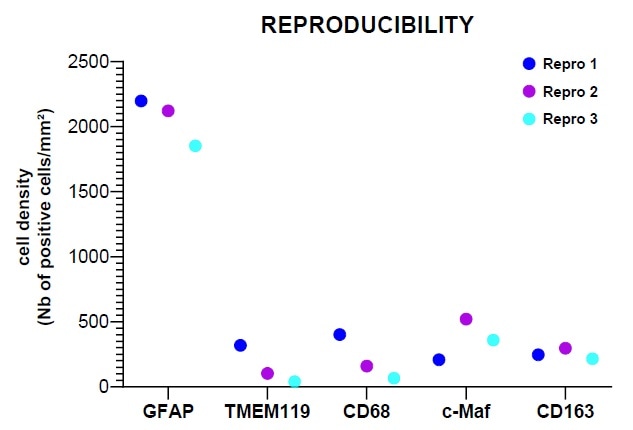
Image Credit: Cerba Research
Figure E highlights that analysis with HALO® showed satisfactory results in terms of the panel’s performance.
HISTOPROFILE® Neuro M1/M2: Infiltration analysis
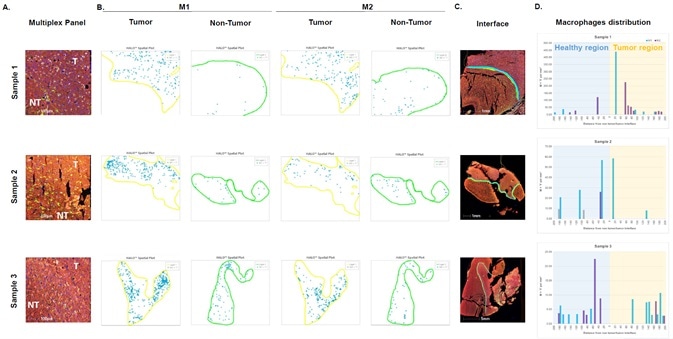
Image Credit: Cerba Research
Infiltrating macrophages were also analyzed at the tumor/non-tumor interface.
Figure A shows a merged image at the interface, highlighting the interface of the tumor (T) and non-tumor (NT) regions using a yellow trace.
Figure B displays HALO® spatial plots which highlight M1 (left) and M2 (right) cells at the interface of the tumor (yellow region) and the non-tumor (green region). The blue circles in the image help visualize phenotyped populations within the landscape.
Figure C shows the region included in the analysis, highlighting this with a line at the interface.
Figure D provides infiltration analysis results, representing the density of M1 (blue bars) and M2 (purple bars) at the interface of the healthy to tumor regions. This highlights the variability of the macrophage distribution at the tumor/non-tumor interface.
Conclusion
The study presented here investigated human GBM tissues using a multiplex panel comprised of GFAP, TMEM119, CD68, c-Maf and CD163, the HISTOPROFILE® Neuro M1/M2. Using multi-spectral acquisition, it was possible to easily phenotype and localize microglia and macrophage populations in the tissues examined.
The study also quantified microglia and peripherally recruited M1 and M2 macrophages, evaluating the spatial distribution of these populations within the tumor core, margin and edge.
The work showcased here highlights the capabilities of multiplex immunohistochemistry and its ability to phenotype and spatially analyze resident and recruited immune cell populations.
This analysis is possible on a single tissue section, opening up a number of possibilities for the potential application of this method in clinical studies.
Acknowledgments
Produced from materials originally authored by Nicolas Goulange, Amanda Finan-Marchi, Maroua Tliba, Manon Motte, Fanny Estermann, Alexandre Vernay, Jean-Philippe Coton, and Renaud Burrer.
About Cerba Research
For over 35 years, Cerba Research has been setting the industry standard for exemplary clinical trial conduct. Today, across five continents, with a focus on precision medicine, we are changing the paradigm of the central lab’s role in complex clinical research.
From protocol inception through development and to market, our passionate experts deliver the highest quality specialized and personalized laboratory and diagnostic solutions. Partner with us for the most efficient strategy to actualize your biotech and pharmaceutical products sooner and improve the lives of patients worldwide.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.