This article is based on a poster originally authored by Chris Bohl, Ken Brouwer, Karissa Cottier, Scott Heyward, Courtney Noah, Brian Ogilvie, and BioIVT.
Several reports in the literature have documented variations in drug pharmacokinetics and/or pharmacodynamics among subjects in clinical cohorts or target patient populations. Further analysis of these cases has revealed that genetic factors and distinct demographics contribute to these variations.1,2,3,4 In some instances, this has required dosage adjustments for specific patient populations.5
The challenges include the evolving nature of clinical scientific research and the need to incorporate populations historically underrepresented in clinical trials. Additionally, considerations for diversity are often lacking in pre-clinical test systems used during the development of new drug entities, reflecting similar challenges in clinical trials.
It is possible to mitigate donor-dependent variability in pre-clinical test systems, such as cryopreserved primary human hepatocytes and human liver microsomes, by creating pooled test systems from multiple donors. However, it remains unclear whether pooled test systems can accurately predict drug ADME properties across all demographics.
In the studies discussed here, a large-scale evaluation was conducted to assess the impact of incorporating diversity into in vitro ADME/DMPK test systems. This was achieved by leveraging extensive characterization data generated during the production of cryopreserved primary human hepatocytes (n=814) and human liver microsomes (n=653). Notably, enzyme activity data was collected for both Phase I (P450) and Phase II drug-metabolizing enzymes.
Methods
Metabolism enzyme activities
The average metabolic enzyme activity rates, Phase I (P450) and Phase II drug metabolizing enzymes, were determined using historical characterization data acquired during the production of cryopreserved primary human hepatocytes and human liver microsomes.
The validated methods which are typically enclosed on product specific datasheets were used to measure drug metabolizing enzyme rates. Donor data were grouped and evaluated based on donor race including African American, Asian, Caucasian, and Hispanic, each of these groups was stratified further by gender.
Average metabolic enzyme activity was also determined based on age criteria from neonatal infants to older adults and BMI classification, i.e., underweight, healthy, overweight, and obese. The average specific activity of drug-metabolizing enzymes was compared to a miscellaneous control group derived from all donors across all datasets.
The trends observed in specific enzyme activities across various groups and test systems were determined via a threshold of ≥20 % difference from the control average.
Table 1: Donor Demographics. Source: BioIVT
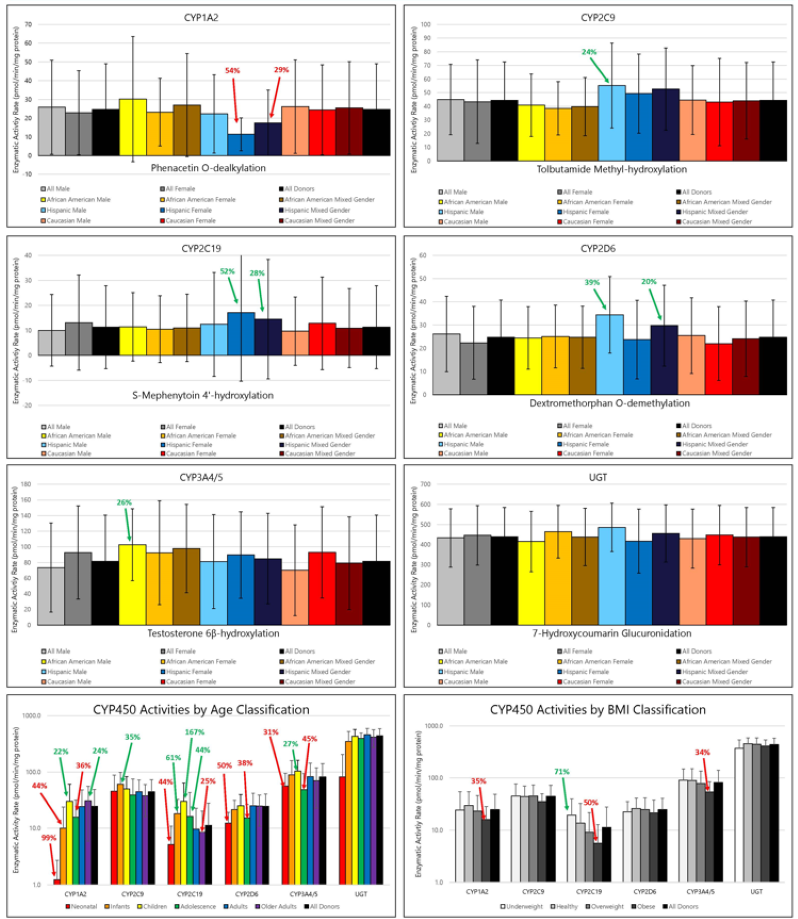
Figure 1. Average Activities of Major Drug Metabolizing Enzymes in Cryopreserved Human Hepatocytes Grouped by Gender and Race, Age or BMI. Image Credit: BioIVT
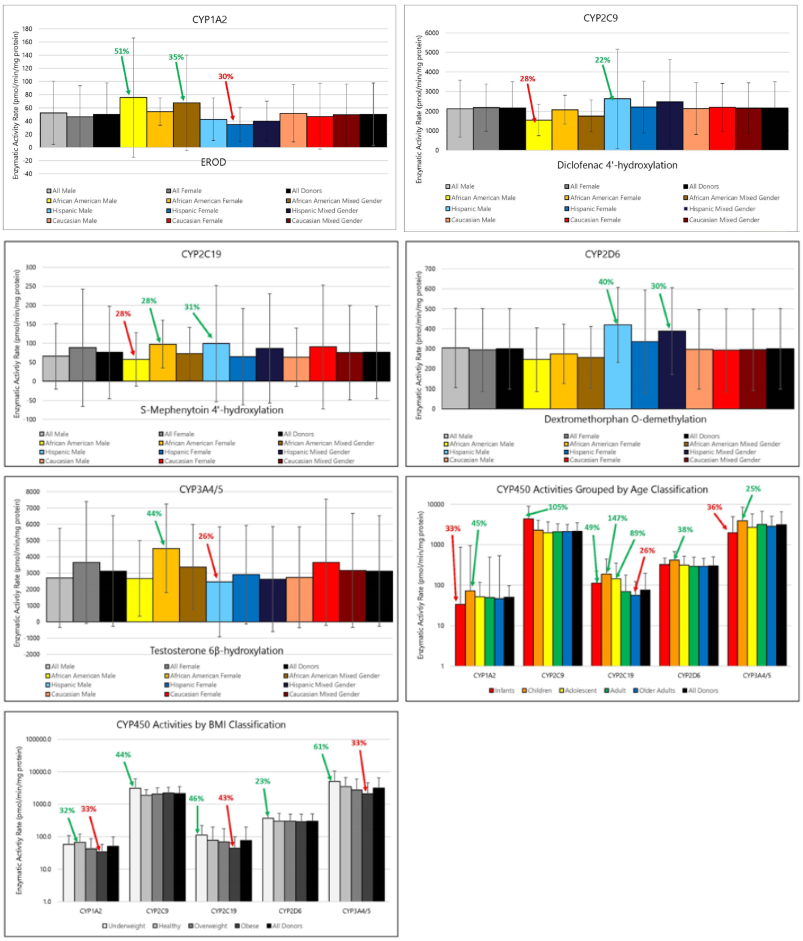
Figure 2. Average Activities of Major Drug Metabolizing Enzymes in Human Liver Microsomes Grouped by Gender and Race, Age or BMI. Image Credit: BioIVT
Conclusions
The lack of diversity in donated tissues used in ADMET test systems is evident, with Caucasians comprising 77-82 % of the datasets. This highlights the need for the development of more diverse human hepatocyte and liver microsome pools, which could enable the validation of experimentally predicted changes in drug-metabolizing activities.
Significant donor variability in metabolic enzyme rates was observed across all enzymes. However, when enzymes such as CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4/5, and UGT were grouped by gender, race, age, and BMI, no significant differences in average activity rates were detected in either human hepatocytes or human liver microsomes.
Additionally, datasets from some underrepresented Asian populations were too small to be included in the study, highlighting the challenges of recruiting and including underrepresented populations in in vitro ADMET test systems. This limitation restricts the full statistical power of the study.
While trends in average enzyme activities were observed across all groups, these trends were not consistently replicated in both hepatocytes and microsomes. Only CYP1A2 and CYP2D6 showed similar activity trends across both systems.
References and further reading
- Guttman Y. et al. Front.Genet (2019)
- Chu T. US Pharm (2014)
- Claudio-Campos K. et al. Drug Metabol Persoanl Ther (2015)
- Bhatt DK. et al. Clin Pharmacol Ther (2019)
- Nguyen AB. et al. Front Cardiovasc. Med (2022)
About BioIVT
BioIVT, formerly BioreclamationIVT, is a leading global provider of high-quality biological specimens and value-added services. We specialize in control and disease state samples including human and animal tissues, cell products, blood and other biofluids. Our unmatched portfolio of clinical specimens directly supports precision medicine research and the effort to improve patient outcomes by coupling comprehensive clinical data with donor samples.
Our Research Services team works collaboratively with clients to provide in vitro hepatic modeling solutions. And as the world’s premier supplier of ADME-Tox model systems, including hepatocytes and subcellular fractions, BioIVT enables scientists to better understand the pharmacokinetics and drug metabolism of newly discovered compounds and the effects on disease processes. By combining our technical expertise, exceptional customer service, and unparalleled access to biological specimens, BioIVT serves the research community as a trusted partner in ELEVATING SCIENCE®.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.