The sudden outbreak of SARS-CoV in China has caused an epidemic situation in 2003. Similar to SARS-CoV-2, the causative pathogen of the currently ongoing coronavirus disease 2019 (COVID-19) pandemic, SARS-CoV belongs to the B lineage of the betacoronavirus family. The genomes of both coronaviruses share 79% sequence similarity. Mechanistically, SARS-CoV initiates the host cell process by interacting with cell membrane receptor ACE2 via the RBD of spike glycoprotein. This is followed by the fusion of the viral envelope with the host cell membrane and the release of viral RNA into the host cytoplasm.
Even after the complete termination of SARS-CoV transmission in July 2003, some zoonotic cases (animal to human transmission) have been identified sporadically. The viral isolates of these cases were different from those observed in the previous outbreak. Moreover, some incidences of laboratory-acquired infections have been identified in some countries. These observations highlight the possibility of future SARS outbreaks and demand for effective therapeutic and preventive interventions.
The study
The scientists have identified a potent antiviral nanobody (S14) from alpacas (South American camelids) that had been immunized with the recombinant spike RBD of SARS-CoV. Moreover, they have investigated the neutralizing potency of S14 against SARS-CoV.
Nanobodies are the smallest single-domain antibodies derived from heavy-chain antibodies found in camelids. Nanobodies have gained immense attention as promising therapeutic agents because of high stability in extreme temperature and pH and the ability to recognize unique epitopes that conventional antibodies cannot bind.
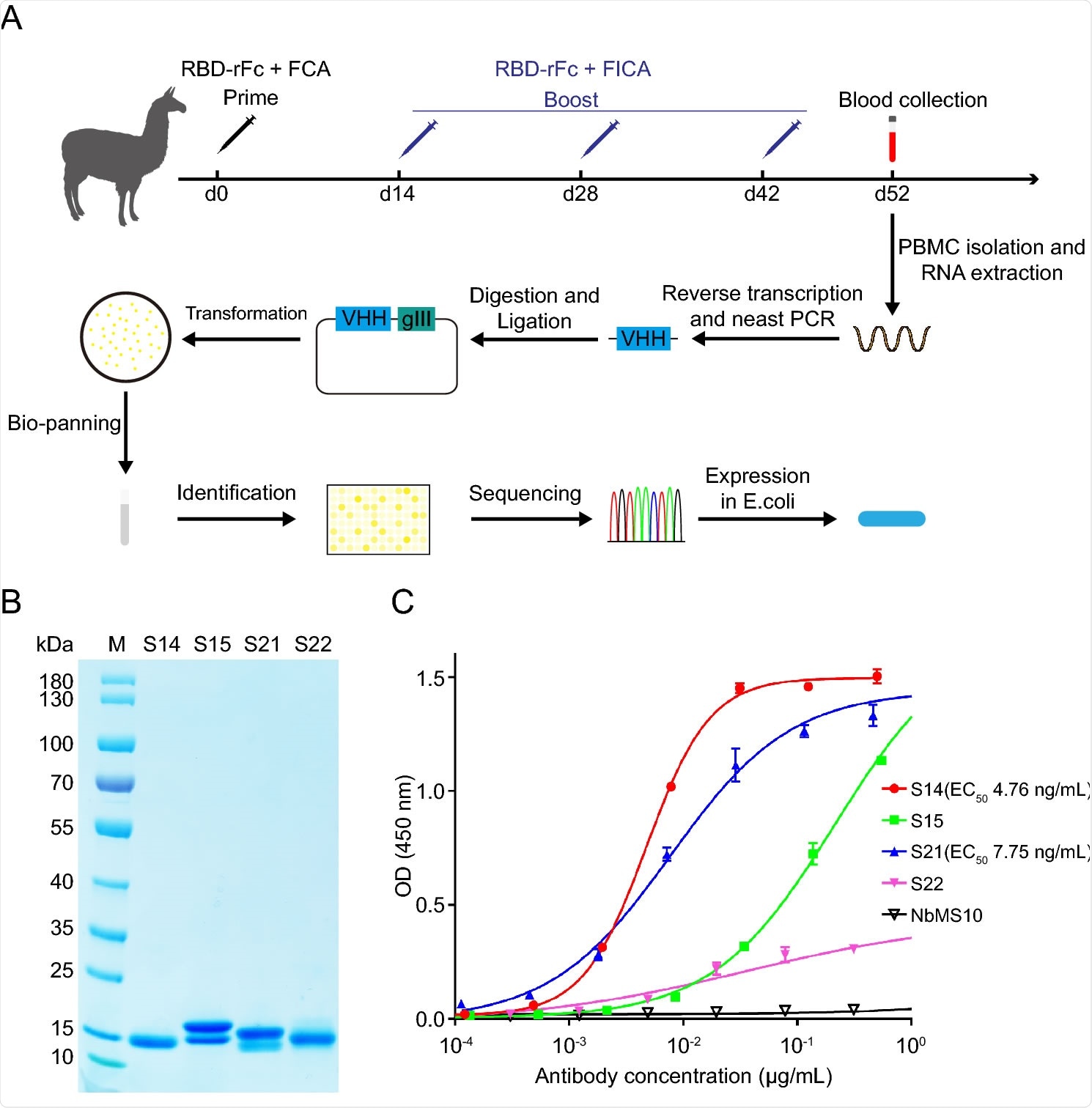
Generation and identification of SARS-CoV-RBD-specific nanobody. A Schematic illustration of the construction of SARS-CoV nanobody library and generation of SARS-CoV-RBD-specific nanobody. RBD-rFc SARS-CoV RBD with rabbit IgG-Fc tag (Sino biological), FCA Freund's complete adjuvant, FICA Freund's incomplete adjuvant, VHH variable region of heavy chain antibody. B SDS-PAGE analysis of purified SARS-CoV nanobodies. M Marker. The molecular weight is indicated on the left. C Evaluation of the binding activity between SARS-CoV RBD and nanobodies. Results are presented as the mean values of optical density at the absorbance of 450 nm ± standard deviation (n = 2).
Preparation of nanobodies against SARS-CoV spike RBD
The scientists used phage-display technology to construct a nanobody library from peripheral blood mononuclear cells isolated from immunized alpacas. In total, they conducted four rounds of bio-panning, followed by identification of monoclonal phages by enzyme-linked immunosorbent assay (ELISA) and sequencing. Finally, they selected four positive clones (S14, S15, S21, and S22) to purify nanobodies using affinity chromatography.
Characterization of anti-SARS-CoV nanobodies
The scientists performed ELISA to characterize selected nanobodies. The findings revealed that three out of four nanobodies bind efficiently with SARS-CoV RBD. Based on these findings, they selected S14 nanobody for further characterization.
They performed bio-layer interferometry and pseudovirus neutralization assay to determine the binding kinetics and neutralization efficiency of S14 against SARS-CoV, respectively. The findings revealed that S14 binds to SARS-CoV RBD with high affinity and efficiently blocks the entry of SARS-CoV into host cells.
In contrast, S14 did not exhibit any binding or neutralizing activity against SARS-CoV-2 pseudovirus, indicating high specificity of S14 against SARS-CoV.
Mechanism of virus neutralization
The scientists performed competitive binding assays and flow cytometry to determine how S14 nanobody neutralizes SARS-CoV. The findings revealed a dose-dependent inhibition of SARS-CoV RBD – soluble ACE2 interaction by S14 nanobody. Moreover, inhibition of SARS-CoV RBD – cell-expressed ACE2 interaction was observed at high concentrations of S14 nanobody. However, this inhibitory activity of S14 was abolished at low concentrations.
According to available literature, the residues Y442, L472, N479, D480, and T487 present at the SARS-CoV RBD – ACE2 interface play vital roles in host receptor adaptations, cross-species infections, and evolution of SARS-CoV. Given this observation, the scientists generated a set of RBD mutants, including Y442A, L472A, N479A, D480A, and T487A, to determine whether these residues are involved in S14-mediated neutralization.
The findings revealed that S14 nanobody efficiently binds to all RBD mutants, indicating that the virus neutralization potency of S14 is not affected by these mutations.
Study significance
The study identifies a high-affinity nanobody targeting the spike RBD of SARS-CoV. Furthermore, this nanobody efficiently and specifically blocks the interaction of SARS-CoV RBD with both soluble and cell-expressed ACE2. Based on these observations, the scientists believe that the nanobody can be used as a potential therapeutic agent against SARS-CoV infection.
Source