Taylor dispersion analysis, also referred to as TDA, is an orthogonal method used for stability and sizing analyses of biomolecules in solution. Research has shown that hydrodynamic radius (Rh) measurements obtained from TDA on monoclonal antibodies (mAbs) and other mono-disperse, unstressed protein-based systems are analogous to data acquired using dynamic light scattering (DLS), a well-known, complementary method. Although both these techniques are capable of detecting differences in oligomeric state and self-association between samples, they react differently depending on which mixtures are present.
TDA and DLS Techniques
The DLS technique is highly sensitive when large species are present. However, this method tends to hide data regarding the smaller species in the sample. TDA measurements in mixtures, on the other hand, are capable of resolving the smaller species in the sample and are not considerably influenced when trace quantities of aggregates or larger species are present.
Insulin is a key bio-therapeutic used for treating diabetes mellitus. It presents a useful case study for assessing the application of the TDA method for defining the stability and hydrodynamic radius (Rh) of protein-based biopharmaceuticals present in solution, and also for emphasizing variations in the data acquired for oligomeric mixtures from DLS measurements.
Insulin oligomers are known to dissociate at low protein concentrations at neutral pH; this is an important feature that is integral to protein’s function in the body. The more stable form of insulin is hexameric insulin which is used to store the protein inside cells, and the more active form of insulin is monomeric insulin, which adheres to insulin receptors.
As a result, it is essential to detect the insulin’s oligomeric state in a particular formulation so as to estimate the formulation’s stability. When insulin monomers undergo self-association, this results in dimers which in turn join to form hexamers. It is possible to observe the changes in the oligomeric state of insulin through their change in size, where smaller sizes relate to oligomers of lower order.
TDA serves as a useful technique to establish the hydrodynamic size. This is achieved by tracking the dispersal of a nanoliter volume sample pulse as it passes via a microcapillary. As a result of a combination of diffusion and dispersion, the sample pulse widens at a certain level by virtue of the solute molecules’ hydrodynamic size. The time-evolved concentration profile, also known as Taylorgram, determined with UV absorption at a particular wavelength is tracked at dual permanent points along the microcapillary. Upon examination of the Taylorgrams, the molecular diffusion coefficient and thus the solute molecules’ hydrodynamic radius can be easily measured.
The section below describes the measurements performed to track the oligomeric state and self-association of insulin present in the solution with reducing concentration at neutral pH.
Experimental Framework
First, insulin provided by Sigma Aldrich was prepared in a PBS buffer and then filtered. Next, TDA measurements were carried out on a Malvern Panalytical Viscosizer at a temperature of 20°C with an uncoated capillary and a 214nm wavelength filter. Throughout the measurement process, default measurement settings for sizing were used. Then, DLS measurements were carried out on a Malvern Panalytical Zetasizer Nano at 20°C temperature. Finally, Thermo Scientific’s NanoDrop 2000 was used to determine the concentrations.
Results and Discussion
Figure 1 shows example Taylorgrams for monomeric insulin and hexameric insulin samples, indicating variations in peak width with hydrodynamic radius. This data can be used to establish the molecule’s hydrodynamic radius. Molecules having a smaller hydrodynamic radius have narrower peak widths, while molecules having a larger hydrodynamic radius exhibit broader peaks. Variations in hydrodynamic radius can be used in order to identify a change in the self-association state of a molecule.
The DLS information reveals that the hydrodynamic radius of insulin in PBS does not show any changes, within the limits of experimental ambiguity. The TDA data, on the other hand, displays a distinct change in the insulin’s hydrodynamic radius as well as a clear trend with concentration. When insulin’s concentration reduces, the recorded size reduces considerably, thereby suggesting a decrease in the oligomeric state of insulin. Figure 2 depicts the hydrodynamic radius (Rh) in PBS for a range of insulin concentrations as determined by both techniques.
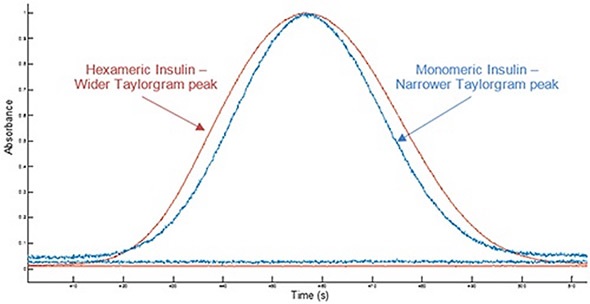
Figure 1. Overlay of example Taylorgrams (UV absorbance vs time) from Viscosizer measurements for samples of hexameric insulin and monomeric Insulin (note Absorbance normalized to 1).
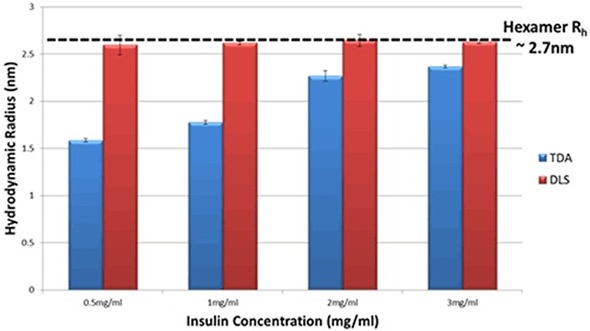
Figure 2. Hydrodynamic Radius measurements for a series of different insulin concentrations in PBS buffer at 20°C as determined by TDA and DLS.
In order to verify a size reduction with insulin concentration in PBS, the samples were evaluated by means of size exclusion chromatography or SEC. Data obtained from the SEC technique demonstrates an increase in retention volume as the insulin concentration reduces, thereby showing a decrease in the size of insulin. This ambiguity in the data occurs because DLS is mainly an intensity-weighted method and hence sizes viewed by this technique are biased towards the size of the largest oligomer found in the sample, which is an insulin hexamer in this case. In contrast, TDA together with UV detection is a mass-weighted method and hence provides significant measurement advantages when it comes to detecting small differences to the target molecule under consideration.
As indicated in the data for insulin solutions (Figure 2), the use of comparable information from TDA can provide a better understanding of the molecules’ self-association state. This is because measurements are not significantly influenced when trace quantities of aggregates or larger species are present in the solution.
Conclusion
In this article, insulin was taken as a case study, which showed how TDA can be applied for tracking size variations caused by changes in a protein’s oligomeric state, along with a concentration change. The study also demonstrated how TDA can be used to give complementary size data to present-day methods like DLS, thanks to its innate ability to monitor variations in oligomeric state without any major bias from aggregates or larger species in the solution.
References
- A. Hawe, W.L. Hulse, W. Jiskoot, R.T. Forbes, ‘Taylor Dispersion Analysis compared to Dynamic Light Scattering for the size analysis of therapeutic peptides and proteins and their aggregates’ Pharm Res 28: 2302-2310 (2011)
- A. Lavoisier, J-M Schlaeppi, ‘Early developability screen of therapeutic antibody candidates using Taylor Dispersion Analysis and UV area imaging detection’ mAbs 7:1, 77-83 (2015)
- A.M. Gualandi-Signorini, G. Giorgi, ‘Insulin formulations - a review’ European Review for Medical and Pharmacological Sciences 5: 73-83 (2001)
- Malvern Panalytical White Paper ‘Understanding Taylor Dispersion Analysis’ www.malvern.com
About Malvern Panalytical

Malvern Panalytical provides the materials and biophysical characterization technology and expertise that enable scientists and engineers to understand and control the properties of dispersed systems.
These systems range from proteins and polymers in solution, particle and nanoparticle suspensions and emulsions, through to sprays and aerosols, industrial bulk powders and high concentration slurries.
Used at all stages of research, development and manufacturing, Malvern Panalytical’s materials characterization instruments provide critical information that helps accelerate research and product development, enhance and maintain product quality and optimize process efficiency.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.