To arrive at the particle size distribution of samples in a liquid suspension, nanoparticle tracking analysis (NTA) uses the characteristics of both Brownian motion and light scattering. A laser beam is transmitted via a sample chamber and the suspended particles in the beam path disperse light in such a way that is possible to observe clearly using a camera-mounted 20x magnification microscope.
The camera’s frame rate is about 30fps and it is capable of obtaining a video file of the particle movement
due to Brownian motion. The field of view is about 100x80x10μm (Figure 1).
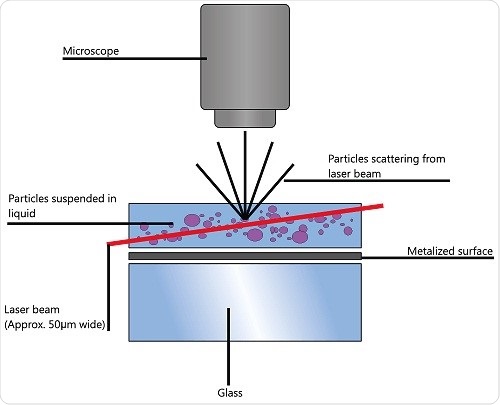
Figure 1. Schematic of the optical configuration used in NTA.
Particle movement is obtained on a frame-by-frame basis. The center of each observed particle is concurrently determined and monitored using the proprietary NTA software and the average distance traversed by each individual particle in the x and y planes is ascertained. Using this value, the particle diffusion coefficient (Dt) is obtained. From the Dt value obtained and with known temperature T and solvent viscosity η, it is possible to obtain the sphere equivalent hydrodynamic diameter d of the particles with the Stokes-Einstein equation (Equation 1), where KB is Boltzmann’s constant.
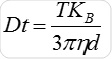
NTA cannot be considered as a collective method involving a huge number of particles, but each particle has a unique size regardless of the neighboring particles. Figure 2 shows the size distribution profile obtained using NTA.
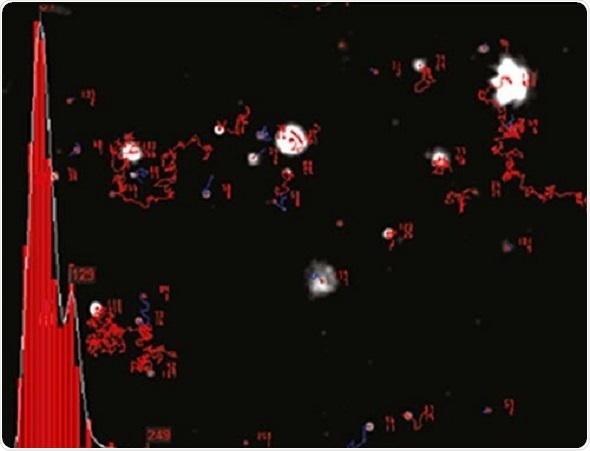
Figure 2. An example of the size distribution profile generated by NTA. The modal size for this sample is found to be approximately 70 nm, with larger sized particles also present.
Since the particles are measured in a known volume the concentration in terms of particles per mL for an overall total or a specific size class can be determined..
Exosome and Microvesicle Characterization
It is important to note that deploying high-intensity laser beams along with a low-background optical configuration enables the observation of deeply sub-micron particles. The lower particle size ranges that can be determined are based on the refractive index of the particles.
Precise size measurement for particles having a very high refractive index like colloidal gold can be obtained down to a diameter of 10nm. However, for particles having a low refractive index as those with a biological origin like exosomes, 30-40nm may be the lowest detectable size.
However, this lower detectable size threshold enables the study of exosomes and microvesicles, whose size is typically far lesser than the 300nm detection limit for many flow cytometers that are available commercially. When a particle’s Brownian motion becomes limited to a diameter of 1-2μm, it can become difficult to track accurately, at this point the upper size limits have been reached.
It is possible to replace the laser that illuminates the nanoparticles with one that helps exciting fluorescence, enabling labeling of the nanoparticles with fluorescent molecules for observation, monitoring, sizing, and concentration measurement with the help of suitable optical filters. As well as providing typical light-scatter data for a sample, all lasers available with NanoSight instruments are capable of exciting fluorescent molecules.
Hence, a 532nm green laser diode can be employed for exciting a variety of organic fluorophores including Nile red and DiI. For individual detection of semiconductor CdSe nanocrystals also called quantum dots, a violet 405nm or blue 488nm laser diode can be used. For the excitation of traditional dyes like those historically utilized in flow cytometry, a 488nm laser diode can be employed.
It is possible to attain phenotyping within complicated mixtures by using fluorophore labeling that is antibody-mediated. Especially significant in this regard is that a specific exosome type can be speciated using Antibody (Ab)-labeling, and also the exosome size can be concurrently determined by studying its Brownian motion. These two measurements are not related to each other. It is also possible to recover the concentrations of these labeled exosomes and compare them against the total number of same-sized structures.
Comparison of NTA to Flow Cytometry and Electron Microscopy
In NTA, nanoparticle size is measured by determining the particle behavior during dynamic Brownian motion, which is not based on the scattering light quantity. However, in flow cytometry, size estimates depend on the light intensity dispersed by a particle normally at a low angle. Hence, for precise measurements, pre-calibrations with particles having almost the same refractive index as the sample nanoparticles are required. Otherwise, there may be a need for sufficient knowledge of the sample nanoparticles pertaining to their light-scattering characteristics.
Several studies were performed by a number of researchers over the years regarding the deployment of NTA in different applications. The first attempts to design an integrated technique that involves fluorescence NTA and flow cytometry for characterization of nanovesicles and microvesicles was completed by Dragovic et al. (2011b)
The flow cytometry analysis by Dragovic et al. (2011a) on a human placental vesicle preparation along with an anti-placental alkaline phosphatase antibody (NDOG2-Qdot605) that was fluorophore-labeled showed that out of 93.5% of the vesicles, which were labeled positive for NDOG2, more than 90% of the vesicles were less than 1000nm in diameter. The major population was between 300nm and 400nm in diameter.
However, while analyzing the same sample by fluorescence NTA, a size distribution of NDOG2-labeled vesicles in the range of 50-600 nm was observed from the results having peaks at 100 and 180nm.While studying total cellular vesicles in ultracentrifuge pellets of platelet free plasma (n=10), it was observed that 200-fold more vesicles could be identified using NTA (mean vesicle size 251±35nm) when compared to flow cytometry. From these results, it was shown that NTA has higher sensitivity than traditional flow cytometry, and they are suitable for studying microvesicles and nanovesicles (Dragovic et al. 2011b).
Although flow cytometry has been believed as not being able to routinely determine exosome preparations, Robert et al. (2012) have shown that highly sensitive flow cytometry offers access to standardized small-size microparticle measurement. Baj-Krzyworzeka et al. (2012a) have provided a comprehensive review on the application of flow cytometry for studying exosomes and microparticles.
Current Detection and Analysis Methodologies
A key issue associated with isolating and purifying exosomes from complicated matrices such as bodily fluids is the scarce number of methods available for evaluating fractions for exosomal concentration and content measurement.
According to Van der Pol et al. (2010), in spite of a significant amount of clinical and scientific interest, there are no currently agreed standard methods available for isolating, identifying and characterizing exosomes and microparticles as their size cannot be determined by traditional detection methods like flow cytometry. According to him, different method combinations are required to clinically identify the appropriate characteristics of exosomes and microparticles. However, as bodily fluids are biologically complicated, microvesicle separation remains a huge challenge.
He then suggested that light scattering methods of dynamic light scattering (DLS) and NTA could determine the absolute and relative size distributions of microvesicles in a few minutes. Additionally, Raman spectroscopy could determine the concentration, size and biochemical composition of unlabeled single microvesicles although the time taken for measurement is in hours. From fluorescence-based optical techniques, fluorescence correlation spectroscopy (FCS) and fluorescence NTA (fNTA) could determine the absolute size distribution and acquire biochemical data by the application of fluorescent antibody labeling, though this was a challenge and there were a number of optical and practical issues. For size, concentration, cellular origin of microvesicles at high speed and biochemical composition, fNTA was recognized as most appropriate, particularly because it could directly identify the properties of microvesicles in the body fluid itself.
In the recent past, Müller (2012) has studied the emergence of new tools for analyzing cell-type specific exosomes and microvesicles (EMVs), suggesting several technologies for density, size and molecular composition of EMVs along with techniques for better separation and purification from heterogeneous vesicle populations. He also suggested that the advancements in micro-flow cytometry, AFM, biosensing, mass spectrometry and nanoparticle tracking would enable qualitative and quantitative analysis of all the EMV components. Preferred technologies are those that offer signatures unique for EMV subsets instead of a single or few averaged parameters for the whole EMV population.
New Commercial Tests
With increasing interest in this area, several novel technologies and reagents for the separation, purification, and study of exosomes have been formulated in the recent years, and are also available in the market:
- Exomir™ deploys a unique approach wherein samples are transferred above syringe filters for exosome and bigger membrane-bound particle manufacture, then rinsed using an RNA extraction reagent for the lysing of the trapped particles in order to study the same using qPCR.
- Exotest™ is a patented sandwich ELISA kit for the trapping and quantification of exosomes based on a tumor-associated marker, caveolin-1 (Logozzi 2009) and expression of housekeeping proteins (CD63 and Rab-5b) and for identification of exosomes in melanoma patient plasma as a tool for cancer follow-up and screening.
- Exosome Diagnostics Inc. is formulating several molecular diagnostics using binding reagent libraries to develop tumor-specific biomarkers for isolation of exosomes from cancer patients for further investigations by means of traditional sandwich immunoassay methods. The formulations are based on research carried out by Balaj et al. (2011).
- Exosome Display is an innovative technique adopted by Anosys Inc. based on technology designed by Delcayre et al. (2005), for customizing exosomes with the required characteristics and manipulating exosome composition.
- ExoQuick™ is a proprietary exosome precipitation reagent based on polymers, enabling the extraction of protein and one-step microRNA biomarker from exosomes in bodily fluids, such as plasma, for qPCR profiling. NTA was deployed for validating exosome precipitation by this technology (Systembio Technical Manual 2011).
- Caris Life Sciences has developed Carisome™, a diagnostic technology that is blood-based, traps and characterizes circulating microvesicles, even exosomes. It is founded on work initially performed by Skog et al. (2008).
- Separation of exosomes from blood and other bodily fluids is made possible by Exosome Sciences’ 96-well assay, using which researchers can extract exosomes from bodily fluids with their Enzyme Linked Lectin Specific Assay (ELLSA). This, in turn, allows analysis by means of detection molecules like antibodies that are linked to a particular biomarker on the exosome.
- A novel reagent has been lately elaborated by Life Technologies for separating exosomes from biological media and biological fluids to be used along with their RNA marker identification system Ion Torrent (Magdeleno, 2012). Recently, this reagent was given the status of “complete exosome workflow solution: from isolation to identification of the RNA markers using the Ion Torrent Personal Genome Machine” by Vlassov (2012a) using NTA to confirm that their reagent is highly efficient and as good as ultracentrifugation for exosome separation. Also, Zeringer (2012) has elaborated the adoption of this reagent for exosome concentration from a range of sample types for the purpose of downstream analysis.
- 101Bio Inc has developed a PureExo® Exosome Isolation Kit (2013) recently that claims 95% isolation efficiency of intact exosomes in less than 2 hours from plasma or serum without the need for ultra-centrifugation.
- Exo-spin™ is another kit advertised as appropriate for preparing functional, pure exosomes from a range of biological fluids, including saliva, cell culture media, urine and blood plasma/cera. It has been also advertized as offering a high-speed option when compared to ultracentrifugation and a higher efficiency than competitor kits. (Exo-Spin, 2013). NTA was used for validating the product’s quality.
- Norgen’s Urine Exosome RNA Isolation Kit has also been commercialized as a comprehensive system for concentration and separation of exosomal RNA from tissue culture media and RNA. The isolation and purification from urine is performed by using spin column chromatography with the isolation matrix being Norgen’s proprietary resin, subsequent to which lysing of exosomes is performed for RNA release that is next attached to Norgen's resin (BIND) for further analysis.
- exoEasy Maxi Kit® is a recent introduction and is advertised for purification of extracellular vesicles from animal or human plasma/sera or from cell culture supernatants using spin columns to remove contaminating proteins, organic polymers and other impurities in under 30minutes.
- HansaBioMed (2013) offers several products for studying exosomes, such as immunobeads. NTA-analyzed exosome standards are also marketed by the company.
It is important to note here that all the tests are concentrated on exosome separation from complex bodily fluids like urine or blood, to be evaluated further using traditional technologies, such as ELISA and qPCR or other functional assays. As such, there is no opportunity to individually characterize, enumerate and phenotype the exosomes so they could be suggested as bulk purification/separation procedures.
The Emergence and Assessment of NTA as MV Characterization Method
Subsequent to early studies of applying DLS for microparticle measurement (Harrison 2008; Harrison et al. 2009), Gardiner et al. (2009 and 2010) began deploying NTA for visualizing and for concentration and sizing determination of cellular microparticles and exosomes. Other researchers started evaluating NTA for discussing analytical and pre-analytical problems in the analysis of blood microparticles (Yuana et al. 2011) and for concentration measurement and microparticle sizing by means of light scattering methods (Gabriel and Giordano, 2010).
Consequently, Sokolova et al. (2011) described the classification of exosomes resulting from human cells by NTA and SEM, while Dragovic et al. (2011) continued their work to both the sizing and phenotyping of cellular vesicles using NTA. Further studies monitored explicitly on the use of NTA for the analysis and concentration measurement of circulating microparticles (Gardiner, 2011); the analyses of in vivo derived human extracellular vesicles (Taylor, 2011); the analysis of cell exosome and nanovesicle emission (Powis et al. 2011) and the observing of microvesicle and exosome secretion from immune cells (Soo et al. 2012). Cicek Gercel-Taylor et al. (2012) later used NTA in the analysis of circulating cell-derived vesicles in ovarian cancer patients.
The quantification and profiling of exosomes in human plasma using protein microarray was also associated to NTA (Jørgensen et al. 2012) and in the segregation, concentration measurement and classification of exosomes from regular urine (Dimuccio et al. 2012).
Vlassov and his team have studied exosomes, providing an outline about their biological functions, composition, therapeutic and diagnostic potential and have arrived at the following conclusions:
- Exosomes are microvesicles comprise of protein and nucleic acid, which all cells secret.
- An abundance of exosomes is seen in all bodily fluids, such as urine, saliva and blood.
- Exosomes play the remarkable role of intercellular communication.
The composition, functions and pathways of exosomes and their employment in therapeutic and diagnostic applications are also elaborated (Vlassov et al. 2012b). They offer a number of instances of NTA analysis of exosomes in liquid samples. They have also proved how easy it is to obtain size and concentration data using NTA in comparison to EM and DLS.
NTA has been regularly used for studying exosomes and microparticles and the data shared in several studies. Certain studies that have been instrumental in determining the physicochemical nature of the microvesicular structures under study are highlighted here:
- Cantaluppi et al. (2013) provided NTA data, including RT-PCR, western blot, FACS and bioanalyzer for isolating, characterizing and presenting pro-angiogenic activity of microvesicles (MVs) obtained from human pancreatic islets.
- Katsuda et al. (2013) demonstrated that human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes, while studying the accumulation of β-amyloid peptide (Aβ) in the brain affected by Alzheimer's disease (AD). In order to validate the size of purified ADSC#4-derived exosomes at 175nm, they studied virus-mediated NEP gene delivery using NTA and TEM, considering that Neprilysin (NEP) is the highly significant Aβ-degrading enzyme. The Bradford method and NTA were used to ascertain the particle numbers and protein amounts of harvested exosomes.
- Hajj et al. (2013) reported that there was a strange secretion from specific cells of cochaperone stress-inducible protein 1 (STI1) through a heterogeneous population of extracellular vesicles. NTA was used for the characterization of the release of MVs from the RAW264.7 macrophage cell line, subsequent to treatment with the proinflammatory cytokine TNF-a (@ 20 ng/mL).
- Shelke et al. (2013) studied the role of fetal calf serum exosomal RNA in in-vitro experiments using NTA.
- Moggio et al. (2013) used NTA for evaluating the microRNA content of extracellular vesicles from rat’s urine for differentiating between healthy vs. polycystic kidney disease.
- Antone et al. (2013) showed that neutrophil/monocyte microvesicles increased in susceptible subjects due to cigarette smoking.
- Royo et al. (2013) in the quest for a new source of non-invasive disease biomarkers and proving that extracellular vesicles released by hepatocytes also carry RNA, have shown that these vesicles, which are involved in stellate cell activation may become a new source for non-invasive identification of the liver toxicity markers. NTA was deployed for the characterization of extracellular vesicles released in two non-tumoral hepatic models: progenitor cell line acquired from a mouse fetal liver and primary culture of rat hepatocytes.
- Raposo and Stoorvogel (2013) have recently come up with a detailed review on the topic of exosomes, microvesicles and associated structures with particular focus on EV characterization. They have also proposed technologies for the formation, function and targeting of EVs.
References
- Antone L, Hoxha M, Angelici L, DiFrancesco M, Pergoli L, Dolo V, Bertazzi PA, PesatoriAc, Bollati V (2013) Cigarette smoking induces and increase in nuetrophil/monocyte microvesicles in susceptible subjects, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
- Arigi EA., Polanco G, Kharaziha P, Panaretakis T (2012) Prostate Cancer Derived Exosomes, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012.
- Baj-Krzyworzeka M, Baran J, Szatanek R and Siedlar M (2012a) Application of Flow Cytometry in the Studies of Microparticles, http://cdn.intechopen.com/pdfs/37431/InTech-Application of flow cytometry in the studies of microparticles.pdf.
- Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy S L, Breakefield XO and Skog J (2011) Tumor microvesicles contain retrotransposon elements and amplified oncogene sequences, Nature Communications, in press.
- Bio Scientific (2011) http://www.biooscientific.com/
- Carisome Diagnostic Technology (2011) https://www.carislifesciences.com/
- Cantaluppi V; Figliolini F; De Lena M; Beltramo S; Medica D; Biancone L; Segoloni GP; Camussi G (2013) Isolation, Characterization and Pro‐Angiogenic Activity of Microvesicles (MVs) Derived from Human Pancreatic Islets: 1176, 24th International Meeting, Abstracts: Latest Advances in Clinical Islet Transplantation; Transplantation: ,27 November 2012 - Volume 94 - Issue - p 160.
- Delcayre A, Estelles A, Sperinde J, RoulonT, Paz P, Aguilar B, Villanueva J, Khine S and Le Pecq J-B (2005) Exosome Display technology: Applications to the development of new diagnostics and therapeutics Blood Cells, Molecules, and Diseases, Volume 35, Issue 2, September-October 2005, Pages 158-168.
- Dimuccio V, Ranghino A, Camussi G and Bussolati B (2012) Exosome isolation, count and characterization from normal urine, Nephrol. Dial. Transplant. (2012) 27(suppl 2): ii3-ii4 DOI:10.1093/ndt/gfs196, FO010.
- Dragovic RA., Gardiner C, Brooks A S, Tannetta DS, Ferguson DJP, Hole P, Carr RJG, Redman CWG, Harris AL, Dobson PJ, Harrison P, Sargent IL., (2011a) Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking
- Analysis, Nanomedicine: Nanotechnology, Biology and Medicine, DOI:10.1016/j.nano.2011.04.003
- Dragovic RA, Gardiner C, Tannetta D, Hole P, Carr R J G, Redman CW, Harrison P and Sargent IL (2011b) Development of Flow Cytometry and Fluorescence Nanoparticle Tracking Analysis (NTA) to Characterise Cellular Microvesicles and Nanovesicles., FlowcytometryUK, 18th 20th July 2011 Royal York Hotel, York, UK.
- ExoCarta—A database of molecules identified in exosomes (2011)
- ExoSpin (2013)https://www.cellgs.com/items/exosomes/exospin.html
- Exostest (2013) Exosome capture and isolation tools - immunobeads. http://hansabiomed.eu/
- Exosome.com (2011)
- Gabriel D A and Giordano K (2010) Microparticle Sizing and Counting Using Light Scattering Methods, Semin Thromb Hemost 36(8): 824-832
- Gardiner C (2011) Update on Nanoparticle Tracking Analysis; Determination and Characterization of (Circulating) Microparticles; Part C : Novel Methodologies, ISTH Council’s XXIII Congress and 57th Annual SSC, Kyoto, July 23-28, 2011.
- Gardiner C, Dragovic R, Brooks A, Alvarez L, Harrison P and Sargent I (2009) Visualisation, sizing and counting of cellular microparticles and exosomes using Nanoparticle Tracking Analysis, Oxford Biomedical Imaging Festival, St John’s College, University of Oxford, Oxford UK 16th September 2009.
- Gardiner C, Dragovic R, Brooks A, Tannetta D, Redman C, Harrison P, and Sargent I (2010) Nanoparticle Tracking Analysis For The Measurement And Characterisation Of Cellular Microvesicles And Nanovesicles, BSHT and NVTH Joint Symposium, June 23-25th, 2010, NH Hotel Leeuwenhorst, Noordwijkerhout, The Netherlands, submitted.
- Gercel-Taylor C, Atay S, Tullis R H, Kesimer M, Taylor DD (2012) Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients, Analytical Biochemistry, Available online 9 June 2012, http://dx.DOI.org/10.1016/j.ab.2012.06.004
- Gyorgy B, Wright M, Nagy G, Toth K, Polgar A, Zelenak G, Borocz I, Turiak L, Herczeg P, Ledeczi Z, Derfalvi B, Vekey K, Kittel A, Gay S, Falus A, Buzas E (2012a) A novel flow cytometric approach reveals abundant CD8+ T cell derived microvesicles in rheumatoid arthritis synovial fluid samples, Ann Rheum Dis2012;71:A19 DOI:10.1136/annrheumdis-2011-201231.3.
- György B, Szabó TG, Turiák L, Wright M, Herczeg P, Lédeczi Z, Kittel Á, Polgár A, Tóth K, Dérfalvi B, Zelenák G, Böröcz I, Carr B, Nagy G, Vékey K, Gay S, Falus A, Buzás EI (2012b) Improved Flow Cytometric Assessment Reveals Distinct Microvesicle (Cell-Derived Microparticle) Signatures in Joint Diseases.PLoS ONE 7(11):e49726.doi:10.1371/journal.pone.0049726
- Hajj GNM, Arantes CP, Dias MVS, Roffé M, Costa-Silva B, Lopes MH, Porto-Carreiro I, Rabachini T, Lima FR, Beraldo FH, Prado MMA, Linden R, Martins VR (2013) The unconventional secretion of stress-inducible protein 1 by a heterogeneous population of extracellular vesicles, Cellular and Molecular Life Sciences, March 2013, DOI 10.1007/s00018-013-1328-y
- HansaBioMed (2013) https://hansabiomed.eu/shop/index.php?route=common/home
- Harrison P (2008), The Application of Dynamic Light Scattering To Measuring Microparticles, ISTH SSC, Vienna, 5th July 2008.
- Harrison P, Gardiner C (2012) Invisible vesicles swarm within the iceberg, Journal of Thrombosis and Haemostasis, DOI: 10.1111/j.1538-7836.2012.04711.x
- Harrison P, Dragovic R, Albanyan A, Lawrie AS, Murphy M, Sargent I (2009) Application of dynamic light scattering to the measurement of microparticles. Journal of Thrombosis and Haemostasis, Volume 7, Supplement 2: Abstract OCTU-056.
- Huang W-L, Lin C-C, Su W-C (2012) Isolation of tumour associated exosomes from clinical samples using the ultra-filtration method, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012.
- Hutcheson JD, New SEP, Goettsch C, Rogers MA and Aikawa E (2013) Regulated release of macrophage-derived matrix vesicles from lipid raft domains: a role for formation of microcalcifications in vulnerable plaques, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
- Jørgensen M, Bæk R, Søndergaard E, Pedersen S, Kristensen S R, and Varming K(2012) Quantification and Profiling of Exosomes in Human Plasma using Protein Microarray, International Society for Extracellular Vesicles meeting - ISEV 2012, Gothenburg, Sweden, 18th to 21st April 2012.
- Katsuda T, Tsuchiya R, Kosaka N, Yoshioka Y, Takagaki K, Oki K, Takeshita F, Sakai Y, Kuroda M and Ochiya T (2013) Human adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomes, Sci Rep. 2013; 3: 1197. , Published online 2013 February 1. doi: 10.1038/srep01197, PMCID: PMC3561625
- Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, Perdicchio M, Marino M L, Federici C, Lessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L and Fais S (2009) High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS ONE 4(4): e5219. DOI:10.1371/journal.pone.0005219
- Magdeleno S (2012) The complete exosome workflow solution: from isolation to identification of the RNA markers using the Ion Torrent Personal Genome Machine, Molecular Diagnostics Europe 2012 (MDE2012), shouldDexter House, London 10-11th may 2012.
- Moggio A, Harsh D, Sticht C, Camussi G, Bussolati B, Gretz N (2013) Healthy vs polycystic kidney disease: A study on MicroRNA content of extracellular visicles from rats urine, Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
- Müller G (2012) Novel Tools for the Study of Cell Type-Specific Exosomes and Microvesicles, J Bioanal Biomed, Volume 4(4): 046-060 http://dx.doi.org/10.4172/1948-593X.1000063
- Nolte-‘t Hoen ENM, van der Vlist EJ, Aalberts M, Mertens HCH, Bosch BJ, Bartelink W, Mastrobattista E, van Gaal EVB, Stoorvogel W, ArkesteijnG J A and Wauben MHM (2011) Quantitative and qualitative flow cytometric analysis of nano-sized cell-derived membrane vesicles, Nanomedicine: Nanotechnology, Biology and Medicine, In Press, DOI:10.1016/j.nano.2011.09.006
- Norgen (2013) https://norgenbiotek.com/products
- Powis SJ, Soo CY, Zheng Y, Cambell EC and Riches A (2011) Nanoparticle Tracking Analysis of Cell Exosome and Nanovesicle Secretion, Microscopy and Analysis, Sept 2011, 25(6), p7-9 (EU).
- PureExo® Exosome Isolation Kit (2013) http://www.101bio.com/product.php
- Raposo G and Stoorvogel W (2013) Extracellular vesicles: Exosomes, microvesicles, and friends, J Cell Biol, Vol. 200 no. 4, 373-383 , The Rockefeller University Press, doi: 10.1083/jcb.201211138
- Robert S, Lacroix R, Poncelet P, Harhouri K, Bouriche T, Judicone C, Wischhusen J, Arnaud L, Dignat-George F (2012) High-Sensitivity Flow Cytometry Provides Access to Standardized Measurement of Small-Size Microparticles, ATVBAHA.111.244616 Published online before print February 9, 2012, DOI:10.1161
- Royo F, Schlangen K, Palomo L, Gonzalez E, Conde-Vancells J (2013) Transcriptome of Extracellular Vesicles Released by Hepatocytes. PLoS ONE 8(7): e68693. DOI:10.1371
- Shelke GV, Lasser C, Lotvall J (2013) Contribution of Fetal Calf Serum Exosomal RNA in in-vitro experiments Second International Meeting of ISEV 2013, Boston, USA, April 17th-20th, 2013 , Journal of Extracellular Vesicles 2013, 2: 20826 - http://dx.doi.org/10.3402/jev.v2i0.20826
- Kog J, Würdinger T; van Rijn S, Meijer DH, Gainche L; Curry WT, Carter BS; Krichevsky AM (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology 10 (12): 1470–1476. DOI:10.1038/ncb1800
- Sokolova V, Ludwig A.-K, Hornung S, Rotan O, Horn PA , Epple M and Giebel B. (2011) Characterisation of exosomes derived from human cells by Nanoparticle Tracking Analysis and scanning electron microscopy, Colloids and Surfaces, B: Biointerfaces, DOI:10.1016/ j.colsurfb.2011.05.013
- Soo C Y, Song Y, Zheng Y, Campbell E C, Riches A C, Gunn-Moore F and Powis S J. (2012) Nanoparticle Tracking Analysis monitors microvesicle and exosome secretion from immune cells. Immunology, 'Accepted Article', DOI: 10.1111/j.1365-2567.2012.03569.x
- Technical Manual (2011) Analysis of ExoQuick Serum and Urine exosomes using the NanoSight LM10, https://www.systembio.com/.
- Tatischeff, I., Larquet, E., Falcón-Pérez, J., Turpin, P., & Kruglik, S. (2012). Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. Journal Of Extracellular Vesicles, 1. Retrieved from http://www.journalofextracellularvesicles.net/index.php/jev/article/view/19179/24812
- Taylor D D (2011) Nanoparticle tracking analyses of in vivo derived human extracellular vesicles Exosomes and Microvesicles 2011, Wyndham Hotel, Lake Buena Vista, Florida October 15 - 17, 2011.
- Urinary Exosome Protein Database (2009). NHLBI. 2009-05-12. http://dir.nhlbi.nih.gov/papers/lkem/exosome/.
- van der Pol E, Hoekstra A G, Sturk A, Otto C, van Leeuwen TG, and Nieuwland R (2010) Optical and non-optical methods for detection and characterisation of microparticles and exosomes Journal of Thrombosis and Haemostasis, Accepted for publication, DOI: 10.1111/j.1538-7836.2010.04074.x
- van der Pol E, van Gemert M J C, Sturk A, Nieuwland R, van Leeuwen TG (2012) Single versus swarm detection of microparticles and exosomes by flow cytometry, Journal of Thrombosis and Haemostasis, Accepted Article (Accepted, unedited articles published online for future issues) DOI: 10.1111/j.1538-7836.2012.04683.x
- van der Vlist EJ, Arkesteijn GJA, van de Lest CHA, Stoorvogel W, Nolte-'t Hoen ENM, Wauben MHM (2012) CD4+ T cell activation promotes the differential release of distinct populations of nanosized vesicles, Journal of Extracellular Vesicles 2012, 1: 18364 - http://dx.DOI.org/10.3402/jev.v1i0.18364
- Vlassov A (2012a) The complete exosome workflow solution: from isolation to identification of the RNA markers using the Ion Torrent Personal Genome Machine, Exosomes and Microvesicles 2012, Sept 30, Orlando, Fl., USA.
- Vlassov A V, Magdaleno S, Setterquist R and Conrad R (2012b) Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials, Biochimica et Biophysica Acta (BBA) - General Subjects, http://dx.DOI.org/10.1016/j.bbagen.2012.03.017
- Yuana Y; Bertina RM; Osanto S (2011) Pre-analytical and analytical issues in the analysis of blood, Microparticles, Thrombosis and Haemostasis, 105.3
- Zheng Y, Campbell EC, Lucocq J, Riches A, Powis SJ (2012) Monitoring the Rab27 associated exosome pathway using nanoparticle tracking analysis, Experimental Cell Research.
- Zeringer E (2012) Concentration of Exosomes from different sample types for downstream analysis using Total Exosome Isolation reagents, Exosomes and Microvesicles 2012, Sept 30, Orlando, Fl., USA
About Malvern Panalytical

Malvern Panalytical provides the materials and biophysical characterization technology and expertise that enable scientists and engineers to understand and control the properties of dispersed systems.
These systems range from proteins and polymers in solution, particle and nanoparticle suspensions and emulsions, through to sprays and aerosols, industrial bulk powders and high concentration slurries.
Used at all stages of research, development and manufacturing, Malvern Panalytical’s materials characterization instruments provide critical information that helps accelerate research and product development, enhance and maintain product quality and optimize process efficiency.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.