Complexes, such as protein/protein, nucleic acids/nucleic acid, protein/nucleic acid, protein/ligand, nucleic acid/ligand, are often difficult to crystallize even on well-defined biological systems, consuming more time and amount of the sample.
Complex crystallization can be performed successfully if appropriate preliminary characterization of the complex is carried out through biophysical methods. Dynamic light scattering (DLS) is a widely accepted method for investigating the crystallizability of a sample.
For investigating protein/ligand crystallization, methods like DSC thermoflour-based optimization strategy are used. Likewise, isothermal titration calorimetry (ITC) is considered as the “gold-standard” method for studying molecular interactions. This article illustrates the usefulness of ITC method for enhancing the crystallization of complexes.
The ITC method can be considered as a true in-solution technique that is capable of providing the entire binding profile between two molecules through a single experiment.
It helps determining the values of binding affinity (Ka), entropy and enthalpy changes (ΔS and ΔH) and stoichiometry (N) precisely. The ITC technique is more advantageous than other biophysical methods because of the absence of:
- Buffer restrictions
- Lower or upper size limit for macromolecules
- Labeling, especially in structural studies
ITC apparatus has recently been used with new processing methods to acquire all the kinematic details on more diverse systems, from a simple ligand to complex RNA folding. However, the main drawback of the ITC method is that it needs comparatively large quantities of sample for the experiment.
Nevertheless, this shouldn’t be a major limitation for structural biologists working on NMR or X-ray crystallography studies because the quantity of sample required is the same as for the ITC analysis. Moreover, in the ITC method the sample is not altered, therefore, recovery and concentration of the sample is possible for subsequent crystallogenesis experiments.
The proper folding of a nucleic acid or a protein that is used in a complex can be examined through ITC methods. A few examples of ITC-based crystallization techniques that use Microcal™ iTC200 from Malvern Panalytical are presented in the following sections.
Example 1: Crystallization of the HIV-1 RNA Dimerization Initiation Site that is bound to an Aminoglycoside Antibiotic
The HIV-1 genomic RNA dimerization initiation site (DIS) is considered as a highly conserved sequence that bears a stem-loop structure. The self-complementary sequence present in the loop facilitates dimerization of the viral genome through the formation of a loop-loop (kissing-loop) complex in between two DIS hairpins.
There were some unforeseen similarities in the crystal structures of the DIS loop-loop complex and the 16 S ribosomal aminoacyl-tRNA site (A site), a target of aminoglycoside antibiotics.
Consequently, it was demonstrated that HIV-1 DIS RNA also binds aminoglycosides, and so it could be an appealing target for designing new drugs based on HIV-1 genomic RNA. In this example, the role of ITC in facilitating co-crystallization of a complex made of aminoglycoside lividomycin and the HIV-1 DIS RNA kissing-loop complex is illustrated.
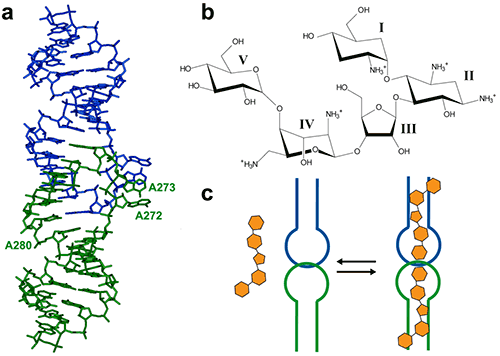
Figure 1. (a) Crystal structure of the HIV-1 genomic RNA dimerization initiation site (DIS) kissing-loop complex (PDB ID 1ZC1, (10)). The two RNA stem-loops are shown in green and blue. Highly conserved adenines 5‘ (A272 and A273) and 3’ (A280) of the 9-nucleotide loop are shown. (b) Chemical structure of the aminoglycoside antibiotic lividomycin. (c) Schematic drawing of lividomycin binding to the DIS kissing-loop complex with a 2:2 stoichiometry.
The interaction of DIS/lividomycin illustrates the salt-dependent binding that is observed often with nucleic acids. Aminoglycosides have a positive charge like most nucleic acids ligands and unspecific interactions caused by electrostatic interactions dominate at low salt concentration.
However, the electrostatic screening at high salt concentrations may interfere with the ligand binding. Thus, in order that unspecific binding is reduced and specific interactions are maintained, preliminary ITC screening is essential for various salt conditions.
Figure 2A shows the results of experiments conducted in a low-salt buffer (25mM KCl, 2mM MgCl2, 25mM Na cacodylate pH 7.0). It can be seen here that a single site model cannot accommodate two binding events.
As expected, at first, a very tight binding is shown with 1:1 stoichiometry when fitted with two independent binding sites model. The second binding site has an affinity in the low micromolar range corresponding to unspecific interactions.
The non-specific binding may happen before a homogenous RNA/ligand complex is formed under crystallization conditions with a concentration greater than 100mM.
However, in case of higher-salt concentrations (200mM KCl, 2mM MgCl2, 25mM Na cacodylate pH 7.0), only specific compact binding is observed, as shown in Figure 2B. Such an experimental set up leads to the formation of RNA/ligand complex, which is suitable for determining crystallization conditions.
Misfolding is a common issue encountered in the structural studies of RNA. In this case, the anticipated stoichiometry is achieved, showing the capability of the ITC experiments to evaluate the appropriate folding of RNAs.
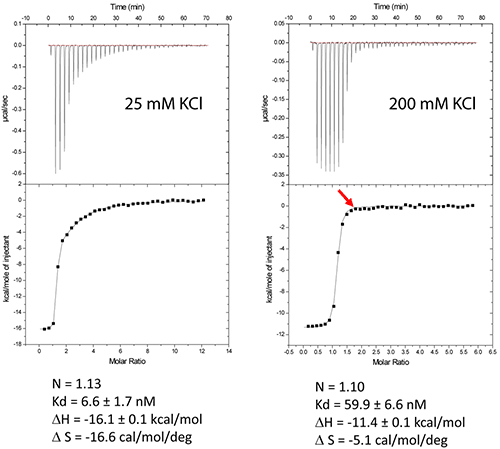
Figure 2. Titration of HIV-1 DIS kissing-loop complex (12 or 6μM RNA in strands) with lividomycin (350 or 400μM) at 25°C in a buffer made with 25mM KCl (left) or 200mM KCl (right), respectively. Data were fitted with a two binding site model (left) or a single set of sites model using Origin analysis module for MicroCal (OriginLab Corp.). Thermodynamic parameters for the specific aminoglycoside binding site are shown.
In ITC-based experiments, it is possible to monitor the stoichiometry in real time, allowing the user to stop the experiment once the required RNA/ligand ration is reached (supposed to be ≥ 1.0). In the titration of DIS RNA/lividomycin at 200mM KCL, the experiment was stopped once the RNA was saturated with the ligand at the 12th injection.
This method should be utilized when the stoichiometry can be determined easily. This is possible when the slope of the sigmoid (Wieseman coefficient c ≥ 100) is steep, and the off-rate constant (koff) is slow.
By following this approach, excess of ligand in the sample can be avoided, and therefore, crystallization can be carried out with an optimal RNA/ligand ratio. A standard example having non-optimized salt concentration is used for the DIS kissing-loop RNA/lividomycin interaction.
A chemically synthesized RNA sequence with a length of 23 nucleotides is used in this example.
The RNA sample is prepared in the ITC buffer up to a concentration of 120µM (in RNA strands) and then placed in the ITC cell. Lividomycin of 2.4mM is taken in the syringe in the ITC buffer. ITC analysis is conducted at a temperature of 12°C for enhanced complex formation and protecting the RNA from potential degradation.
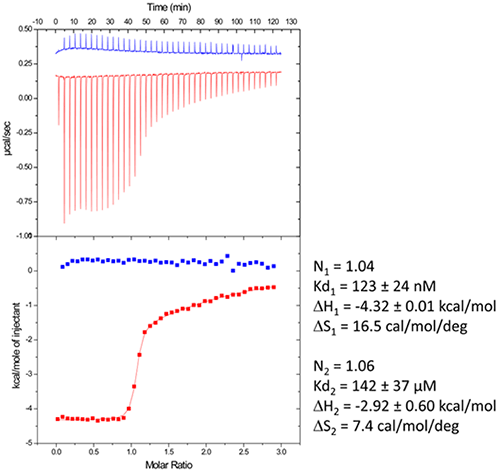
Figure 3. Raw data and binding isotherm for the interaction of [BrU-3] HIV-1 DIS RNA kissing-loop with lividomycin (red). Experiments carried out in MicroCal iTC200. Injection of lividomycin into ITC buffer is shown in blue. Thermodynamic parameters for specific (site 1) and unspecific (site 2) interactions are shown on the right.
After the completion of the ITC experiment, the resulting complex is recovered from the ITC cell and concentrated three folds approximately up to 400µM with the help of microcentrators.
The concentrated solution is then mixed at 1/1 volume in the crystallization trays with the help of a crystallization robot and crystallization sparse matrix screens that are comprised of 96 different crystallization conditions.
The sparse matrix solution and trays are incubated at 37°C inside the crystallization reservoir. After an incubation period of 16-24h under Hampton Research® Natrix HT sparse matrix conditions, large monocrystals ideally suited for diffraction are expected to grow, as shown in Figure 4.
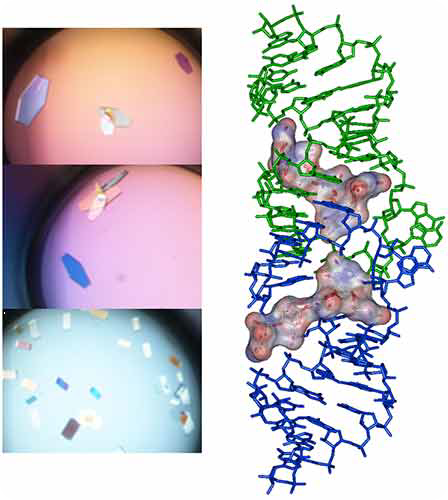
Figure 4. Crystals and X-ray crystal structure of BrU3-DIS kissing-loop complex RNA bound to lividomycin antibiotic (11).
Example II: Crystallization of the E.coli Thiamine Pyrophosphate (TPP) Riboswitch Bound to its Ligand
The untranslated regions of messenger RNA that bind specific metabolites are termed as riboswitches. Once binding occurs, a conformational switch can be observed; this regulates the expression of proteins that cause the biosynthesis of riboswitch substrates.
The E.coli ThiC TPP riboswitch reacts to the coenzyme thiamine pyrophosphate (TPP), which is an active form of vitamin B1, as shown in Figure 5. An 85-nucleotides synthetic RNA fragment that corresponds to the Ecoli TPP aptamer domain is used in this example.
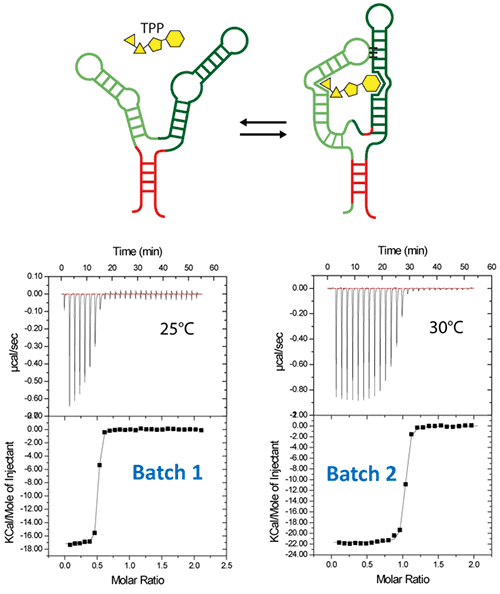
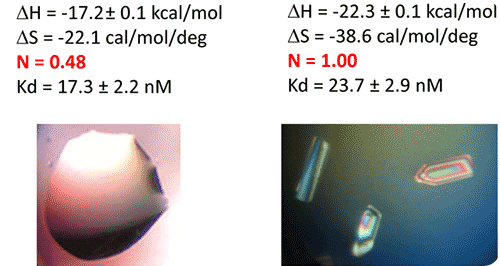
Figure 5. ITC-guided crystallization of the TPP riboswitch bound to its ligand. The secondary structure of the TPP riboswitch is shown on the top (each domain has a different color), the TPP ligand is shown in yellow. ITC experiments performed on a Microcal iTC200 microcalorimeter on RNA Batch 1 and Batch 2 are shown in the middle. The stoichiometry is highlighted in red. The result of crystallization trials is shown on the bottom.
The challenge in using such large RNA with complex secondary and tertiary structures is the retrieval of the folded molecules after overproduction, purification and in vitro renaturation process.
The refolding procedure involves purification, heat denaturation, and flash cooling of the RNA on ice. It is then placed in the ITC buffer (Na cacodylate pH 6.5 50mM, potassium acetate 100 mM, magnesium acetate 5 mM).
After refolding RNA, an ITC experiment was carried out at 25°C. It involved placing 30µM of RNA in the ITC cell, followed by loading the syringe with 500µM TPP. The ITC experiment revealed that only half of the RNA (approx) was able to bind the TPP ligand, which confirms a serious misfolding problem in the RNA, as illustrated in Batch 1 of Figure 5.
The resulting RNA/TPP sample is retrieved from the ITC cell, concentrated further and used for subsequent crystallization trials in the same fashion as the HIV-1 DIS kissing-loop RNA. However, the crystallization assays contain only clear drops, as shown in the left side of Figure 5, which supports the hypothesis that a large part of the RNA is misfolded.
There was a change in the RNA folding protocol in the second step. Following heat denaturation for a period of 3 minutes at 90°C in water, the riboswitch was brought down to room temperature for a period of 40 minutes.
This was followed by the addition of a 10X ITC buffer to a final 1X concentration, and incubation of the mixture at room temperature for a period of 40 minutes. The RNA refolding protocol followed in the second RNA batch (30µM RNA, 500µM TPP) of the ITC experiment resulted in a fully active riboswitch because all of the RNA was able to bind to the TPP ligand.
This is illustrated in Batch 2 of Figure 5. Large monocrystals that diffract up to 5Å resolution were formed subsequent to concentration and introduction of the crystallization drops. The experimental results affirm that ITC technique can be used as a quality control measure in the complex formation before the crystallization trials are conducted.
Example III: Crystallization of the Pseudomonas Aeruginosa Sliding Clamp Bound to Short Peptide Ligands
The DNA polymerase processivity factor or β sliding clamp in bacteria is a homodimer conferring high processivity to DNA polymerases and other factors that are involved in the metabolism of the DNA.
This is achieved by their interaction through a small conserved peptide into a hydrophobic pocket of the clamp. This interaction can be prevented by short synthetic peptides that interfere with the binding of the sliding clamp to the DNA polymerase, and so behave like antibiotics.
This experiment aimed to co-crystallize P.aeruginosa clamp bound to a synthetic peptide to gain structural insights into the interaction and to facilitate structure-based rationale design of novel antibiotic peptides that have a better affinity.
An ITC experiment was performed before crystallization by loading the β sliding clamp (35µM in Hepes pH 7.4 10mM, NaCl 150mM, EDTA 3mM) into the ITC cell and the peptide (300µM) into the syringe. This is shown in Figure 6a.
The complex was used for crystallization trials after increasing its concentration to 47mg/ml with a microcentrator. Crystallization was conducted by mixing equal volumes of the protein/peptide complex with a reservoir solution made with 0.1M sodium Mes pH 6.0 100mM, CaCl2 100mM and 20-30% (w/v) polyethylene glycol (PEG) 400.
The incubation period spanned several days, during which the temperature was maintained at 20°C. At the end of incubation, large monocrystals that diffract up to 2.2Å formed (Figure 6b). Figure 7 shows the X-ray structure.
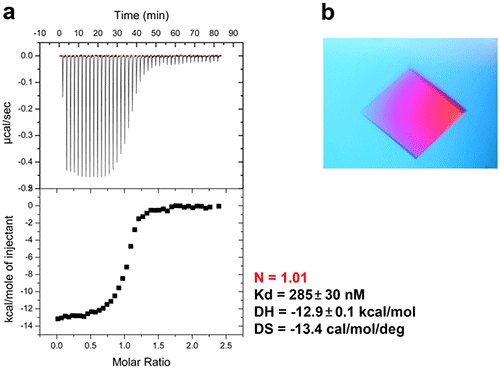
Figure 6. ITC-guided crystallization of the P.aeruginosa β sliding clamp bound to a short peptide. (a) ITC experiment performed at 30°C showing the peptide binding to the homodimeric sliding clamp. (b) Crystal of the peptide/sliding clamp complex obtained from the solution extracted from the ITC cell of MicroCal iTC200.
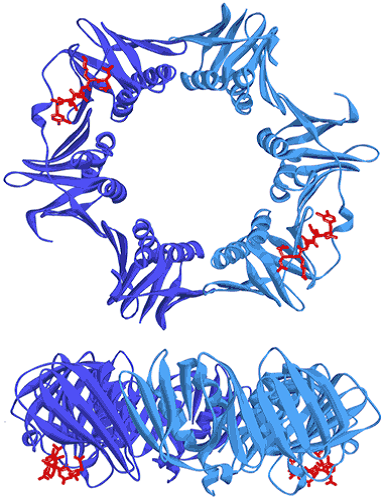
Figure 7. Two views showing the crystal structure of the replicative sliding clamp bound to a peptide with a 2:2 stoichiometry (PDB ID 4TSZ) (16). Protein monomers forming the homodimeric clamp are shown in light and dark blue. Synthetic peptide ligands are shown in red.
Conclusion
The ITC technique can be incorporated into the crystallization workflow of macromolecules complexes, thus increasing the crystallization process’ success rate. The sample requirement in structural studies is apt for ITC analysis.
The real active protein and/or the ligand concentration can be determined by the ITC technique. This technique also aids in optimizing complex formation. With the ITC technique, stoichiometry can be monitored, which helps in determining an optimized ratio of protein and ligand in real time for complex crystallization.
The entire thermodynamic profile of the interaction can also be obtained by simply concentrating the sample subsequent to an ITC analysis.
References
- Ferre-D'Amare, A. R., and Burley, S. K. (1994) Use of dynamic light scattering to assess crystallizability of macromolecules and macromolecular assemblies, Structure 2, 357-359.
- Dupeux, F., Rower, M., Seroul, G., Blot, D., and Marquez, J. A. (2011) A thermal stability assay can help to estimate the crystallization likelihood of biological samples, Acta Crystallogr D Biol Crystallogr 67, 915-919.
- Ladbury, J. E., and Chowdhry, B. Z. (1996) Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions, Chem Biol 3, 791-801.
- Leavitt, S., and Freire, E. (2001) Direct measurement of protein binding energetics by isothermal titration calorimetry, Curr Opin Struct Biol 11, 560-566.
- Privalov, P. L., and Dragan, A. I. (2007) Microcalorimetry of biological macromolecules, Biophys Chem 126, 16-24.
- Burnouf, D., Ennifar, E., Guedich, S., Puffer, B., Hoffmann, G., Bec, G., Disdier, F., Baltzinger, M., and Dumas, P. (2012) kinITC: a new method for obtaining joint thermodynamic and kinetic data by isothermal titration calorimetry, J Am Chem Soc 134, 559-565.
- Ennifar, E., Walter, P., Ehresmann, B., Ehresmann, C., and Dumas, P. (2001) Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site, Nat Struct Biol 8, 1064-1068.
- Ennifar, E., Paillart, J. C., Marquet, R., Ehresmann, B., Ehresmann, C., Dumas, P., and Walter, P. (2003) HIV-1 RNA dimerization initiation site is structurally similar to the ribosomal A site and binds aminoglycoside antibiotics, J Biol Chem 278, 2723-2730.
- Bernacchi, S., Freisz, S., Maechling, C., Spiess, B., Marquet, R., Dumas, P., and Ennifar, E. (2007) Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: thermodynamics and effect on the kissing-loop to duplex conversion, Nucleic Acids Res.
- Ennifar, E., and Dumas, P. (2006) Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies, J Mol Biol 356, 771-782.
- Ennifar, E., Paillart, J. C., Bodlenner, A., Walter, P., Weibel, J. M., Aubertin, A. M., Pale, P., Dumas, P., and Marquet, R. (2006) Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell, Nucleic Acids Res 34, 2328-2339.
- Winkler, W. C., and Breaker, R. R. (2005) Regulation of bacterial gene expression by riboswitches, Annu Rev Microbiol 59, 487-517.
- Winkler, W., Nahvi, A., and Breaker, R. R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression, Nature 419, 952-956.
- Burnouf, D. Y., Olieric, V., Wagner, J., Fujii, S., Reinbolt, J., Fuchs, R. P., and Dumas, P. (2004) Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases, J Mol Biol 335, 1187-1197.
- Wolff, P., Olieric, V., Briand, J. P., Chaloin, O., Dejaegere, A., Dumas, P., Ennifar, E., Guichard, G., Wagner, J., and Burnouf, D. Y. (2011) Structure-based design of short peptide ligands binding onto the E. coli processivity ring, J Med Chem 54, 4627-4637.
- Wolff, P., Amal, I., Olieric, V., Chaloin, O., Gygli, G., Ennifar, E., Lorber, B., Guichard, G., Wagner, J., Dejaegere, A., and Burnouf, D. Y. (2014) Differential modes of peptide binding onto replicative sliding clamps from various bacterial origins, J Med Chem 57, 7565-7576
About Malvern Panalytical

Malvern Panalytical provides the materials and biophysical characterization technology and expertise that enable scientists and engineers to understand and control the properties of dispersed systems.
These systems range from proteins and polymers in solution, particle and nanoparticle suspensions and emulsions, through to sprays and aerosols, industrial bulk powders and high concentration slurries.
Used at all stages of research, development and manufacturing, Malvern Panalytical’s materials characterization instruments provide critical information that helps accelerate research and product development, enhance and maintain product quality and optimize process efficiency.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.