Aggregation of both synthetic and biological materials is a great concern, posing problems at different stages of the discovery and formulation processes. The capability to identify and characterize aggregates is of broad utility, and the experiments discussed in this article demonstrate the ability of the nanoparticle tracking analysis (NTA) technique, that is one of a kind, to comprehend the dynamics of aggregation in a biological system through high resolution particle-by-particle analysis.
A model system, which has been used in drug discovery research and presents some unique challenges in the characterization of reagents, was used in the experiment described in this article. This system was generated to enable functional assembly of recombinant/cloned membrane receptors on liposomes, without the need for reconstituting full length receptors.
The aim of this study was to correlate the earlier published activity data1,2 with the changes in size and aggregation state of the liposome-RTK complex determined by NTA. The experiments described here are expected to represent the characterization methods that are highly useful to the nanoparticle field.
Determining the size, assessing the polydispersity, and identifying the components that promote aggregation are all of interest and can be assessed through these simple experiments.
Background to liposomes
For some time now, drug discovery and drug delivery have used liposomes, and the biophysical characterization of these systems and their payloads is essential in order understand their function. The membrane-associated receptor tyrosine kinase (RTK) target is one such payload that is investigated here, and is the major focus of numerous discovery campaigns.
Generally, RTKs are single-pass transmembrane signaling proteins that are hard to purify intact, and are significant targets of several disease pathways. Due to the abundance of activity data available, this model characterization system was selected in this study.
In the work described here, the aggregation state of a representative RTK target (kinase domain of insulin receptor) was investigated first. Then, the RTK was analyzed on an engineered cell-like scaffold, specifically a liposome functionalized with nickel-chelated head groups (described in references 1 and 2).
The nickel head groups facilitate template-directed self-assembly of His-tagged RTKs for a brief period of time such that the kinase domains (cytoplasmic fragments) are oriented as they would be within a cell (Figure 1).
In spite of the understanding that manganese is not a physiologically-relevant counter-ion, magnesium (Mg) or manganese (Mn) was used as the available counter-ion in the assay buffer during the analysis, in order to figure out why Mn might cause an increase in the activity level of kinases. This was done to get a better understanding of the effects of counter-ions using NTA, and to relate this with the activity data that was obtained previously.
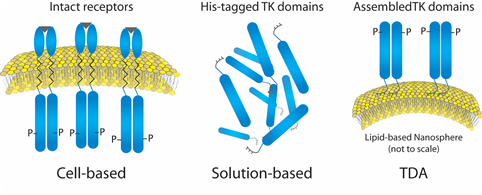
Figure 1. Liposome- Insulin Receptor Complexes.
With the NanoSight range of instruments offered by Malvern Panalytical, this well-studied drug discovery system can be characterized by directly visualizing nanoscale particles in suspension (between 10nm and 2000nm) in real-time with high-resolution and minimal sample preparation.
The NTA technique applies a particle-by-particle approach to particle sizing, providing an unprecedented insight into size distributions. Visual validation and high resolution analysis of particles is provided by NanoSight, within the nanoscale range with both concentration and size measurements. NTA provides a robust analysis of this liposome-protein system and gives an insight into this kinase activation method.
Materials and methods
Blue Sky BioServices (Worcester, MA, USA) provided the purified protein and extruded liposomes. HEPES buffer was used to dilute the sample prior to analysis by a NanoSight NS300 fitted with a 50mW green 532nm laser, a high sensitivity Hamamatsu sCMOS camera, and a 20x objective lens.
NTA 3.1 Build 46 software was used for analyzing samples. NanoSight technology observes the Brownian motion of particles in suspension, and using the Stokes-Einstein equation calculates their size. It observes particles on a frame-by-frame basis through the use of the sCMOS camera (25 frames/second), deriving concentration. The average number of particles per frame in the field of view is determined by the software. This software also calculates a number-weighted concentration result.
Results and discussion
Using NTA afforded an appreciation of the rapid kinetics with high resolution analysis of aggregation on a real-time basis. There was a simultaneous decrease in concentration and an increase in size over time in the presence of Mn buffer.
As is evident in Figures 2 and 4, the liposomes aggregate in the presence of Mn. Analyzing the insulin receptor RTK domain in HEPES (or Tris) buffer indicate that they further aggregate in the presence of Mn (Figures 3 and 4).
This analysis was difficult as the protein itself appears aggregated, to an extent, in the absence of either counter-ion when noting the initial time curve in both Figures 3 and 4, though there is a considerable increase in aggregate size in the presence of Mn that is not observed when Mg is added.
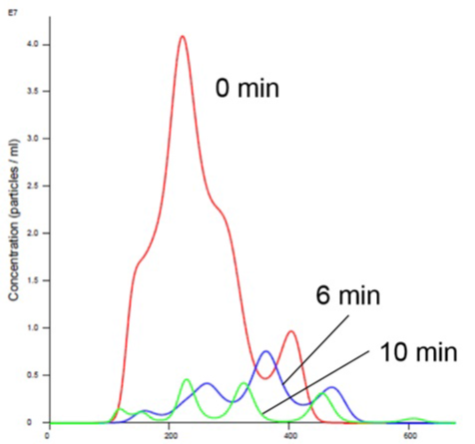
Figure 2. Liposomes in Mn Buffer show aggregation (increased size and decreased concentration).
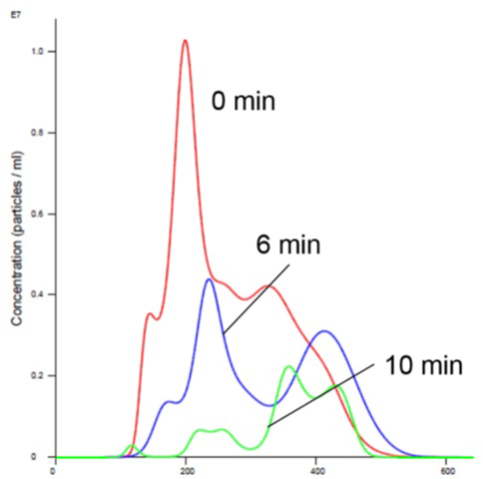
Figure 3. Insulin receptor alone in Mn buffer showing aggregation (increased size and decreased concentration).
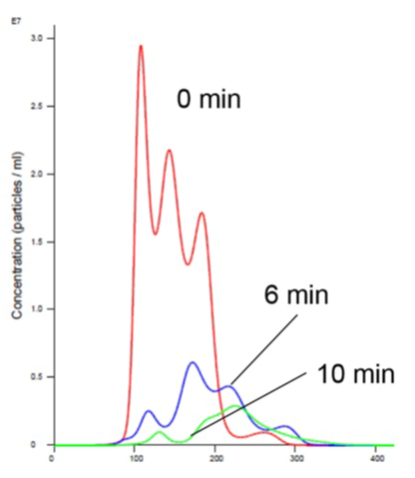
Figure 4. Insulin receptor and liposomes in Mn buffer showing aggregation (increased size and decreased concentration).
Adding Mg to RTK does not promote aggregation and is consistent with the previously-generated data which does not show any appreciable activity in the presence of Mg1,2,5 (Figures 5 and 6).
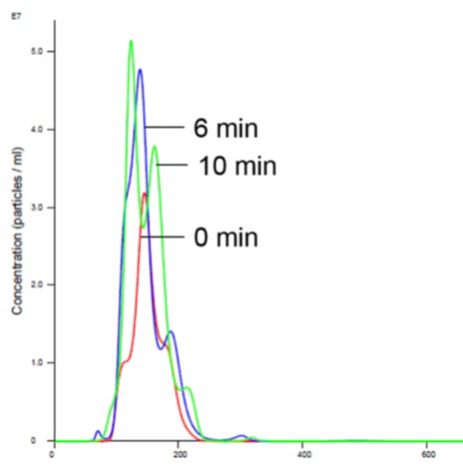
Figure 5. Liposomes in Mg buffer show stabilized size and concentration.
The hypothesis that Mg is a biologically relevant counter-ion but that it does not result in any activity is confusing. The addition of Mg to RTK and liposomes did not cause aggregation but does result in high cell like activity1,2,5. RTK is thought to assemble on the outside of the liposome in a similar manner as inside the cell (Figure 1).
Adding Mn promotes aggregation and increases activity in a simple and cost-effective manner, with considerable robustness and reproducibility. When RTK is properly self-assembled on a liposome in the presence of a biologically relevant counter-ion – Mg, the following benefits are observed 1,2,5:
- increase in autophosphorylation and substrate phosphorylation
- improved substrate selectivity
- highest catalytic rates (except where autophosphorylation is self-inhibitory, Tie2)
As the target seems to be in cell-like conformation, which allows a unique opportunity to screen protein-protein interactions, template-assembled enzymes are expected to be a better screening tool.
This article emphasizes the need for better understanding of the difference between functional assembly of RTK complexes (for interesting colocalization discussion, see reference 4) and forced colocalization (or molecular crowding). The work discussed here is consistent with the hypothesis that Mn facilitates the formation of aggregated complexes, and so increases the catalytic activity of some kinases.
Conclusion
NTA is a valuable method for the accurate characterization of the aggregation behavior of liposome preparations under a variety of conditions. The high-resolution data offers a better understanding of the aggregation kinetics of these systems.
The results presented here question the relevance of compounds selected in high throughput screening (HTS) assays using Mn as a counter-ion.
References
- Esposito EA, Shrout AL, Weis RM. J Biomol Screen 2008; 13 (8): 810-6.
- Shrout AL, Esposito EA, Weirs RM. Chem Biol Drug Des 2008; 71 (3): 278–281.
- Langmuir, 2007. 23(6): p. 3280-9.
- Curr Opin Struct Biol, 2006. 16(6): p. 702-9.
- Gridley S, Shrout AL, Esposito EA, Prog. Mol. Bio. Transl. Sci. 2010; 91:209-39 TDA technology used to prepare the liposomes is protected by patent and available via Blue Sky Bioservices for license: US: 8,541,190.
About Malvern Panalytical

Malvern Panalytical provides the materials and biophysical characterization technology and expertise that enable scientists and engineers to understand and control the properties of dispersed systems.
These systems range from proteins and polymers in solution, particle and nanoparticle suspensions and emulsions, through to sprays and aerosols, industrial bulk powders and high concentration slurries.
Used at all stages of research, development and manufacturing, Malvern Panalytical’s materials characterization instruments provide critical information that helps accelerate research and product development, enhance and maintain product quality and optimize process efficiency.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.