The biological function of proteins relies on their ability to form structured, ordered complexes such as homo-oligomers or hetero-oligomers. Attributes such as stability, activity, and localization can be controlled by these complexes.
Gaining insights into the formation and controlling mechanism of these complexes is essential to understand protein signaling pathways and their control over physiological responses better. The assembly of virulence factors complexes secreted by pathogenic microbes is a subject of intense research.
These factors aid infection, colonization, and survival of the host, thus making them critical for the onset of diseases. The potential to target and suppress these virulence factors paves the way for the development of advanced anti-microbial therapies.
However, it is necessary to gain knowledge about the formation mechanism of homo- and hetero-oligomers. The combination of sophisticated light scattering detectors and liquid chromatography techniques is used for characterizing the protein-protein interactions of specific microbial virulence factors.
Materials and Methods
Proteins and materials were prepared by a procedure outlined elsewhere. SEC-TD, consisting of a triple detector array coupled with a size exclusion chromatography (SEC) system, TDA (Malvern Panalytical, UK), was used to make molecular mass and intrinsic viscosity measurements. The TDA consisted of a fixed light scattering cell equipped with two photodiode detectors, at 90° for right angle laser light scattering (RALS) and at 7° for low angle (LALS); a differential viscometer; a photometer; and a differential refractive index detector.
Both the refractive index detector and photometer were used to determine the protein concentration. The molecular mass M was derived from the concentration, LALS and RALS data. The differential viscometer was used to calculate the intrinsic viscosity (η). The OmniSEC software was used in the calculation of both the intrinsic viscosity and molecular mass.
Experimental Results
This experiment studied the formation and conformation of protein complexes of bacterial virulence factors. The SEC-TD provided key information on the behavior of these proteins.
Formation of Homo-Oligomers
Many factors control the formation of homo-oligomers. The experiment studied the presence of regulating peptides and the effect of calcium binding. Most virulence factors secreted by pathogenic microbes contain the “Repeat in Toxin” (RTX) motif. The calcium response is brought out by the calcium-dependent adenylate cyclase toxin from Bordetella pertussis (CyaA) using the RTX repeat domain (RD). Figure 1 shows the characterization of two RD subdomains, RCS and RCL, for their response to Ca2+ binding.
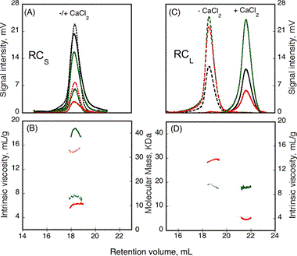
Figure 1. RCS and RCL polypeptide hydrodynamic properties, analyzed using the SEC-TDA method. RCS (A) and RCL (C) polypeptides, without (dashed line) or with (solid line) 5mm CaCl2. The green traces relate to the right angle light scattering; the black traces to the differential refractometer, and red traces to the differential viscometer. RCS (B) and RCL (D) show the intrinsic viscosity (red lines) and molecular mass (green lines) in the presence of (solid lines), or without (dashed lines) calcium.
The RALS (green traces) and the differential refractometer data (black traces) revealed that the molecular weight of the RCL construct is the same in both Apo (- Ca2+) and Holo (+ Ca2+) forms. However, it can be concluded the hydrodynamic volume of the peptide is smaller when attached to calcium from the variations in the retention volumes of 18mL and 22mL for Apo and Holo forms, respectively. Moreover, the intrinsic viscosity is decreased from 13.7 to 3.9mL.g-1 due to binding of calcium.
Conversely, for RCS, the molecular weight is increased from 15.1kDa to 42kDa and the intrinsic viscosity is reduced from 14.7 to 6.4 mL.g-1 due to binding of calcium, but there is no change in the retention volume. These data specify a smaller, closely formed monomeric conformation for RCL, but a tightly structured trimer for RCS with a size same as the unfolded monomer seen in the absence of calcium.
The ATP-dependent proteolytic complex ClpP2 is considered to be helpful in providing the ability of Mycobacterium tuberculosis to stay inactive in host cell phagosomes for several years. In vivo, a tetradecameric complex is formed by ClpP2 for full peptidase activity, but the formation and controlling mechanism of this complex is yet to be known. A disordered N-terminal peptide sequence is thought to influence regulation.
Therefore, SEC coupled to triple detection was used to analyze wild type ClpP2 and an N-terminal truncated form, ClpP2R13 (Figure 2). ClpP2 of 140kDa molecular weight eluted as a single main peak, corresponding to a combination of heptamers and hexamers. Nevertheless, three main peaks were observed at 482kDa, 148 kDa and 46 kDa when the N-terminal prosequence is removed.
The largest molecular weight species is in correspondence with a 21-mer homo-oligomer. This is larger when compared to the tetradecamer expected, thus indicating that the prosequence of ClpP2 regulates the formation of higher order oligomers.
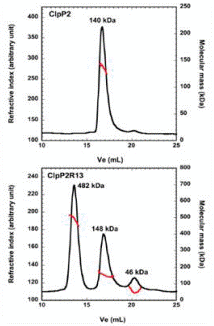
Figure 2. Effect of the removal of a putative propeptide on the production of ClpP2. The ClpP2R13 variant (lower panel) and ClpP2 (upper panel) were analyzed by size exclusion chromatography with triple detection. The black line shows the refractive index, while the red line showed the molecular mass, plotted as an elution volume function. The average molecular weights attained by fixed light scattering are specified at the apex of each peak.
Formation of Hetero-Oligomers
The binding of various regulator proteins, either inhibitors or activators, controls the activity of many proteins. The SEC-TD was used to analyze the complex of bacterial virulence factors with particular regulating proteins. Bordetella pertussis contributes to whooping cough and its key virulence factor is CyaA. CyaA influences the formation of pathogenic levels of cAMP subsequent to the binding and activation by Calmodulin (CaM).
Figure 3 shows the SEC-TD data of the hetero-complex and the individual components. The CaM-CyaA catalytic domain (AC) complex’ molecular weight was found to be 58.4kDa, which roughly equivalent to the sum of the molecular weight of the individual components. This indicates that the complex comprises CaM and AC at a stoichiometry ratio of 1:1.
This was further concluded by Analytical UltraCentrifugation (AUC) data. The intrinsic viscosity was found to be 5.2, 4.9 and 5.0 mL.g-1 for AC, CaM and CaM-AC complex, respectively. The CaM-AC complex has a low value, suggesting a strong compaction of AC and CaM upon formation of the complex. Its hydrodynamic radius, as identified by AUC, was consistently 3.3nm instead of the expected 3.7nm. These data indicates a signaling pathway towards the formation of a compact 1:1 heterodimer upon binding of CaM and CyaA.
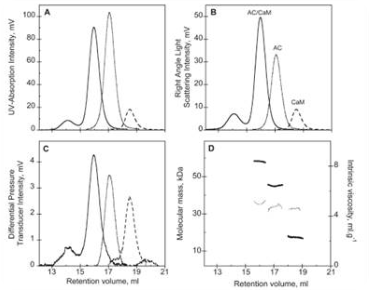
Figure 3. Size exclusion chromatography as followed by a triple detector array of AC (…), CaM (---), and the AC-CaM complex (-): (A) UV absorption chromatogram, (B) right angle light scattering chromatogram, (C) differential pressure chromatogram, and (D) molecular mass (left y axis, thick line) and intrinsic viscosity (right y axis, thin line) for each species.
The micro-organism Klebsiella oxytoca, PulD’s membrane secretin is a component of a multimeric membrane complex needed for effective secretion of virulence factors to the host. PulD combines with the chaperone protein PulS for exact localization of the bacterial membrane. The SEC-TD was used to study the interaction between PulS and the S domain of PulD (Sdom) (Figure 4).
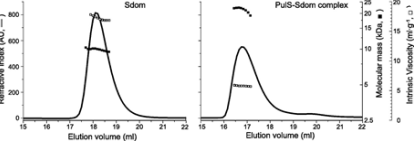
Figure 4. Hydrodynamic properties of Sdom (left panel) and PulS-Sdom complex (right panel) analyzed by SEC-TD, showing refractive index (solid line, left axis), molecular mass (solid squares, inside right axis) and intrinsic viscosity (open squares, outside right axis).
The formation of a Sdom:PulS 1:1 complex with a molecular weight of 22.1kDa was confirmed by the molecular weight measurements. The intrinsic viscosity for Sdom and the Sdom:PulS complex was found to be 16.9 and 5.9 mL.g-1, respectively. These data indicate the formation of Sdom as an elongated disordered structure and the Sdom:PulS complex as an ordered and compacted structure. The hydrodynamic radii values for PulS, Sdom, and Sdom:PulS complex were found to be 1.7 and 2.5, and 2.7nm, respectively. These data also further support the suggestion.
Conclusion
Protein signaling pathways are typically complex due to the involvement of various components and multi-protein signaling complexes. Gaining knowledge about the formation mechanism of homo- and hetero-oligomers is crucial to gaining insights into the activity and regulation of such complexes.
The data presented in this article has demonstrated the compelling data provided by the SEC-TD systems. Such data, including the size and molecular weight of components and complexes, as well as information on protein structure, helps understand the attributes and mechanisms behind these multi-protein signaling pathways.
References
- Karst et al (2010). Calmodulin-Induced Conformational and Hydrodynamic Changes in the Catalytic Domain of Bordetella pertussis Adenylate Cyclase Toxin. Biochem, (2010) Vol.49, p.318-328.
- Sotomayor Perez et al (2011). Calcium-Induced Folding of Intrinsically Disordered Repeat-in-Toxin (RTX) Motifs via Changes of Protein Charges and Oligomerization States. J. Biol. Chem. Vol. 286, p.16997 – 17004.
- Nickerson et al (2011). Outer Membrane Targeting of Secretin PulD Protein Relies on Disordered Domain Recognition by a Dedicated Chaperone. J. Biol. Chem. (2011) Vol. 286, p.38833 – 38843.
- Benaroudj et al (2011). Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2. BMC Biochem (2011) Vol.12, Issue 61.
About Malvern Panalytical

Malvern Panalytical provides the materials and biophysical characterization technology and expertise that enable scientists and engineers to understand and control the properties of dispersed systems.
These systems range from proteins and polymers in solution, particle and nanoparticle suspensions and emulsions, through to sprays and aerosols, industrial bulk powders and high concentration slurries.
Used at all stages of research, development and manufacturing, Malvern Panalytical’s materials characterization instruments provide critical information that helps accelerate research and product development, enhance and maintain product quality and optimize process efficiency.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.