Sponsored Content by AbcamFeb 5 2019
The terminal stages of programmed cell death (apoptosis) can be identified by the condensation of chromatin and fragmentation of the genomic DNA, alongside blebbing of the membrane.
Chromatin Condensation in Apoptotic Cells
As apoptosis occurs, chromatin within the cell undergoes significant structural changes from a heterogeneous network, which is genetically functioning, into a highly condensed, amorphous form.
Condensed chromatin appears brighter than functioning chromatin when stained with nucleotide binding dyes, meaning apoptotic cells can be identified either qualitatively, using fluorescence microscopy, or quantitatively, using flow cytometry.
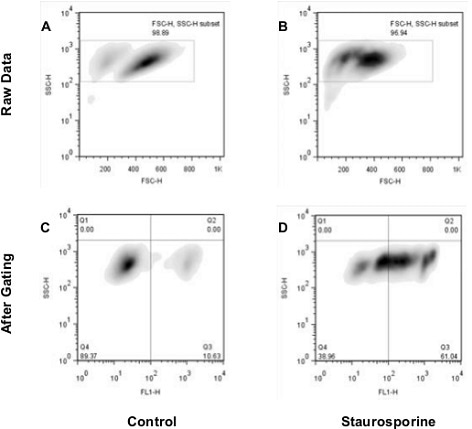
Nuclear Condensation Assay Kit (ab139479): Flow cytometry analysis of untreated staurosporine-treated Jurkat cells. The raw data shown in panels A and B was used for gating cell populations (rectangles) for analysis. Panels C and D represent the separation of healthy and apoptotic nuclei based on their characteristic fluorescence.
Genomic DNA Fragmentation
A nuclease, Caspase-Activated DNase (CAD), is used by cells to fragment and break down the condensed chromatine. CAD is activated by the caspase cascade, which results in DNA cleavage at the sites between nucleosomes, known as internucleosomal linker sites, resulting in the generation of DNA ladders - fragments of around 200 base pairs in size.
An established method of DNA ladder detection is the analysis of the fragmented DNA on agarose gel using an electrophoretic method. This method provides reliable semi-quantitative data and is simple to run.
Some useful information to make sure the research is as effective as possible follow:
- The gel should be ran at a lower voltage than usual as this accounts for the time required for heating and band deformation to occur
- The gel should have a concentration of 1.8 – 2% - this high concentration will give a high enough resolution to see the different steps in the ladder
- The sample DNA loading buffer should be used at a higher concentration than normal, as this will ensure that the sticky, viscous DNA sample remains in the well. E.g. for 50 µL, 10 µL 10X loading buffer should be used
Another method for the detection of DNA fragmentation involves identifying strand breaks/nicks with a TUNEL assay.
The TUNEL technique (short for TdT dUTP nick end labeling) uses terminal deoxynucleotidyl transferase (TdT) to label the ends of nicked DNA strands that are created by CAD. TdT creates a deoxyuridine label on the 3’ blunt ends of the DNA strands
Observing Increases in subG1 Population
DNA fragmentation also results in the number of cells stalled at the G1 stage of the cell cycle, and this can be detected using flow cytometry.
The cell membranes can be permeabilized (e.g. using 70% ethanol), resulting in the leakage of the DNA fragments and cells with a lower DNA content. Following staining using a dye such as PI the G1, S and GM2 cell cycle stages can be profiled using flow cytometry.
The apoptotic cells are clearly found as the high subG1 population located to the left-hand side of the G1 peak. However, this parameter alone is not a perfect indicator for apoptosis as the subG1 fraction contains all dead cells, irrespective of their cell type.
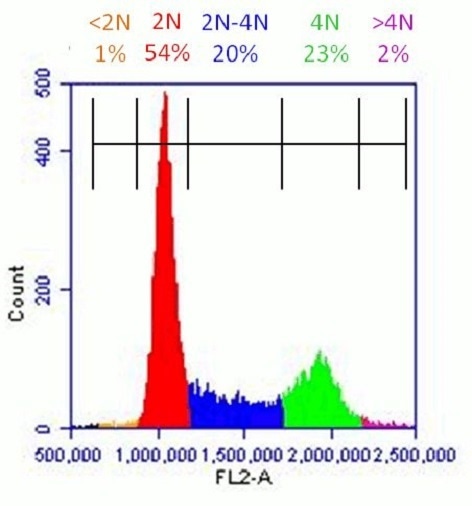
Healthy population of HeLa cells analyzed with Propidium Iodide Flow Cytometry Kit (ab139418): Flow cytometer software was used to establish markers on a histogram plot to quantify the percentage of cells with <2N (dead cells), 2N (G1 phase), 2N-4N (S-phase), 4N (G2 phase) and >4N (M phase) content.
Membrane Blebbing
Alongside the fragmentation of DNA apoptosis’ final stages involve the blebbing of the dynamic membrane and contraction of cells. During this stage the cytoskeleton of the cell begins to disintegrate, which results in some of the membrane protruding outwards. These protrusions then separate from the cell, removing some of the cell’s cytoplasm with them, to form apoptotic bodies.
The blebbing of the membrane can be seen in live cells with phase-contrast microscopy. If using live cells is not possible, or if cells have been prepared for other research (e.g. chromatin condensations or the quantification of fragmentation) it is possible instead to detect the caspase substrates which are associated with membrane blebbing.
However, as this is not a direct method this could provide incorrect results. Examples of caspase substrates that are released upon membrane blebbing are given below:
- PAK (P21-activated kinase)
- ROCK-1 kinase
- Gelsolin
 About Abcam
About Abcam
Abcam is a global life sciences company providing highly validated antibodies and other binders and assays to the research and clinical communities to help advance the understanding of biology and causes of disease.
Abcam’s mission is to serve life scientists to help them achieve their mission faster by listening to their needs, continuously innovating and improving and by giving them the tools, data and experience they want. Abcam’s ambition is to become the most influential life science company for researchers worldwide.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.