Drug-induced liver injury (DILI) has been a significant issue for pharmaceutical companies, clinicians, and regulatory bodies for a long time. As a broad definition, DILI refers to the adverse drug reactions that can result in serious liver damage and, in some quite severe cases, organ failure and mortality.
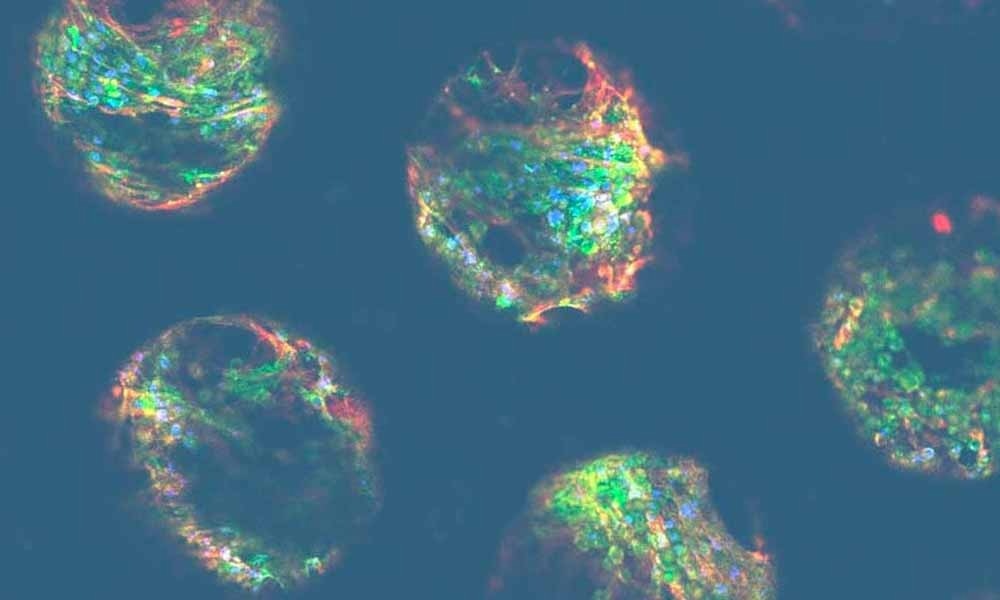
Image Credit: CN Bio Innovations Limited
These adverse reactions are divided into two sub-categories:
- Intrinsic DILI: This is where a drug and/or drug metabolites induce liver toxicity in a predicted and dose-dependent manner. Most individuals show a response at some dose, and the toxicity levels are generally consistent across species. Hepatotoxicity can occur within days, and pathophysiology is similar across incidences.
- Idiosyncratic DILI: On the other hand, drug-induced hepatotoxicity in this category is often unpredictable and does not seem to correlate with dosing. Onset is also rather capricious and can vary from days to weeks to even months. The presenting pathophysiology and mechanism of toxicity differ between people, and only a small number of those exposed to the compound tend to experience these adverse effects. However, what is most concerning is that idiosyncratic DILI is also largely inconsistent across species as well.
It is common knowledge that a majority of drug candidates fail at some stage of the drug development pipeline. DILI is not only responsible for a large proportion of these failures but also causes FDA warnings and drug withdrawal from the market most frequently.
Unfortunately, failure is an unavoidable part of this process; and keeping that in mind, we can agree that it is better for a drug to fail early in the development pipeline, before it is administered to humans; be it through clinical trials or after a widespread release. Having a drug candidate fail earlier in the development process means that stakeholders can reduce the level of investment they put into unsuccessful projects, circumventing the ethical dilemmas concerning human trials, and limiting negative market fallout due to a loss in public trust.
Intrinsic DILI is usually observed during preclinical testing, in the worst-case scenario, it surfaces during the early stages of clinical trials. Idiosyncratic DILI, however, can go undetected until Phase III/IV. This is mainly because:
- Premarketing clinical trials are often underpowered, making them incapable of picking up idiosyncratic DILI in the small group of individuals it would occur in.
- Idiosyncratic DILI also appears to be species-specific. Thus, preclinical models can fail to detect human toxicity.
As expected, idiosyncratic DILI has been a key point of interest for pharma researchers for many years, in a bid to predict the unpredictable. An increase in understanding has also resulted in further sub-categorization of idiosyncratic DILI. In this article, we will focus on immune-mediated toxicity, and more specifically immune-mediated DILI.
Current in vivo approaches struggle to capture the immune component of imDILI
While the classification boundaries are still up for debate, immune-mediated DILI (imDILI) typically refers to a drug-induced immune response against the liver that causes organ damage.
A prototypical example of intrinsic DILI is acetaminophen. Liver metabolism breaks down acetaminophen molecules to create a reactive species that, when over-accumulated (as seen in an overdose), results in hepatocyte necrosis and mitochondrial damage. This acute liver injury is exacerbated when further damage is caused by an immune response, where danger-associated cytokines are released from the necrotic liver cells.
When discussing immune-mediated DILI, we focus on DILI resulting from adverse immune responses, minus the predictable drug-induced hepatotoxicity.
Animal in vivo models of intrinsic DILI are usually quite standard, a large dose of hepatotoxic compounds results in liver damage in most animals. However, there are still complications and caveats which limit the translational relevance of such models. Acetaminophen, for example, shows hepatotoxicity in mice that resembles that of humans. However, rats are much more resistant and need a significantly higher dosing regimen to reach the same toxicity.
Taking these factors into consideration, you can already see that in vivo animal models, as expected, are far less suitable for producing clear imDILI estimations. This is predominantly due to the unique complexities that are found in the human immune system, which are not naturally recapitulated in other species. For example, trovafloxacin and diclofenac are two drugs widely known for their immune-mediated DILI risks, but they do not induce DILI in mouse models.
In an attempt to make mouse models more human-relevant, mice are frequently pre-, co-, or post-treated with inflammagens (such as lipopolysaccharide) to activate an artificial immune response, in the presence of the drug. While this does produce a DILI phenotype, its relevance to humans is questionable for many reasons - the main one being that the toxicity observed is neutrophil-mediated, which is an atypical pathophysiology for human imDILI.
The mechanism underlying this species-dependent disparity in DILI is still under investigation. However, there are some drugs, which produce a human-specific imDILI response, whose mechanisms are better understood. Lumiracoxib and ximelagatran were withdrawn from the market by the FDA after liver damage was reported in a small group of patients. These drugs triggered an adverse immune response through their interaction with specific isotypes of human leukocyte antigens (HLAs), a class of cell-surface proteins that regulate immune-mediated cell destruction.
The HLA complex is unique to humans. While animals do have a similar system called the major histocompatibility complex, they are not identical. To observe immune-mediated DILI in animals, they must be genetically modified to express the HLA isotypes of interest. Having to make these adjustments, not only raises the cost of the in vivo models significantly but also necessitates prior knowledge of which HLAs (a class that comprises many) would be of interest.
Animal models are helpful for screening for intrinsic DILI, offering valuable information. However, they are ultimately unable to recapitulate the uniquely human immune microenvironment; a fact that is underscored by their frequently contradictory results when investigating the mechanisms of idiosyncratic DILI. The inherent complexities associated with whole animal studies also add a layer of difficulty in understanding what is and is not translatable to the clinic.
How do new drug modalities complicate the situation?
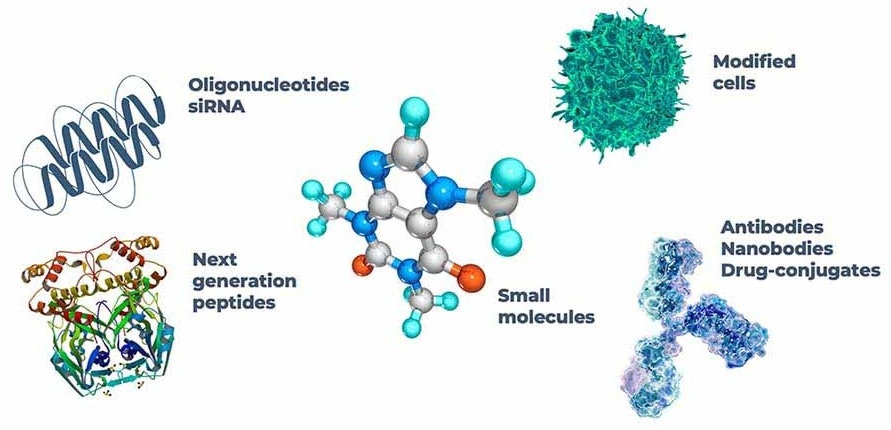
Image Credit: CN Bio Innovations Limited
The evaluation of how effective current models are for drug development extends beyond the world of small molecules. Chemically synthesized and well-characterized, small molecules frequently influence only certain cellular processes. In contrast, newer modalities, such as ‘biologics,’ have considerable variation in both size and composition and work on a much wider scale.
Newer modalities are often tailored to humans. A mouse liver is homologous to a human liver, but there are crucial differences. Several of their enzymatic mechanisms are “like enough” to be comparably affected by small molecules, however, a therapeutic antibody designed specifically to target a human protein will likely experience poor target binding in the same mouse model. Gene-based drugs meet the same fate, as antisense oligonucleotides (ASOs) and small-interfering RNA (siRNA) control gene expression by binding specific RNA sequences.
Some preclinical trials investigating these novel modalities use non-human in vivo models with the assumption that the general pharmacokinetic profile and toxicity are independent of species, even though the target is human-specific. For instance, rat, mouse, and monkey models were employed during toxicological studies of the ASO drug - mipomersen. There were minimal increases in hepatotoxic markers, resulting in FDA approval in 2013, but with a block box warning for hepatoxicity risks. The drug was eventually withdrawn from the market, in 2019, because of these dangers.
As mentioned, genetically engineered animals offer a path forward for incorporating human-specific components into preclinical in vivo models. Computer modeling is another method applied to try and predict the potential adverse effects gene-based therapeutics may have on a person’s unique genetic sequence.
Whilst helpful, both methods require prior knowledge of where adverse reactions might come from, and neither approach assists with predicting the unpredictable. Ideally, alternative models that use human cells and allow for adjustments to be made at a granular level need to be incorporated to study cell-cell interactions to complement, or in time, replace the need for in vivo results.
Traditional in vitro approaches have certain limitations too
How do we use human cells without putting human lives at risk? The solution lies in advanced in vitro models. Numerous drug development pipelines already employ human in vitro models in their preclinical studies, but these fail to flag potential immune-mediated DILI risks because of the absence of ‘adjustable complexity,’ as mentioned above.
Typical in vitro screening methods include hepatocyte cell lines seeded into 2D culture plates. Three areas of improvement exist that are crucial for engineering more predictive in vitro platforms.
- Cell Source: Primary hepatocytes are accepted as the gold standard for human-relevant liver models. They are limited only by their unstable differentiation in 2D. Immortalized cell lines, derived from hepatocellular carcinomas, can easily be obtained and cultured to produce a more stable phenotype. However, due to their reduced hepatocytic functionality and divergent genetics, they can be unsuitable for imDILI assays. In addition, imDILI only occurs in a small subsection of the population meaning multiple donors with diverse genetics are required to detect imDILI activity in such studies.
- Microenvironment: Signals from the extracellular matrix (ECM), alongside autocrine, juxtacrine, and paracrine communication are all vital for building in vitro liver tissue, with functionality that can be compared to the in vivo organ. By adding cytokines exogenously, or coating the tissue culture plastic with an ECM, we can reproduce the different molecular cues needed in a 2D culture plate, but a 3D culture needs to capture the incredibly important temporal and spatial aspects of these signals.
- Co-cultures: Whilst it is important to utilize relevant cells and culture them in a suitable 3D environment, imDILI models must include immune components, such as adaptive (T-cells) and/or innate (Kupffer cells) immune systems to be useful as an in vitro imDILI screening tool.
Incorporating these improvements no doubt necessitates the utilization of novel platforms in place of standard 2D culture plates. Growing primary liver cells in 3D increases the culture’s sensitivity to hepatotoxic compounds, and it substantially enhances the longevity of the cells. This allows for longer and repeat dosing regimens, which more closely resemble what is observed in the clinic.
Recently, scientists have used 3D methodologies to generate hepatocyte cellular spheroids. These clusters of cells self-assemble into spherical structures that express a functional morphology that closely resembles their in vivo counterparts, especially when compared to equivalent 2D cultures. While this is an improvement, these types of cultures are restricted by their static environment. It is also important to note that the liver is a highly vascularized organ, with metabolically active hepatocytes. So, active cell culture perfusion is needed to imitate the hepatic physiochemical environment appropriately.
Immune-competent Organ-on-a-chip platforms address these issues
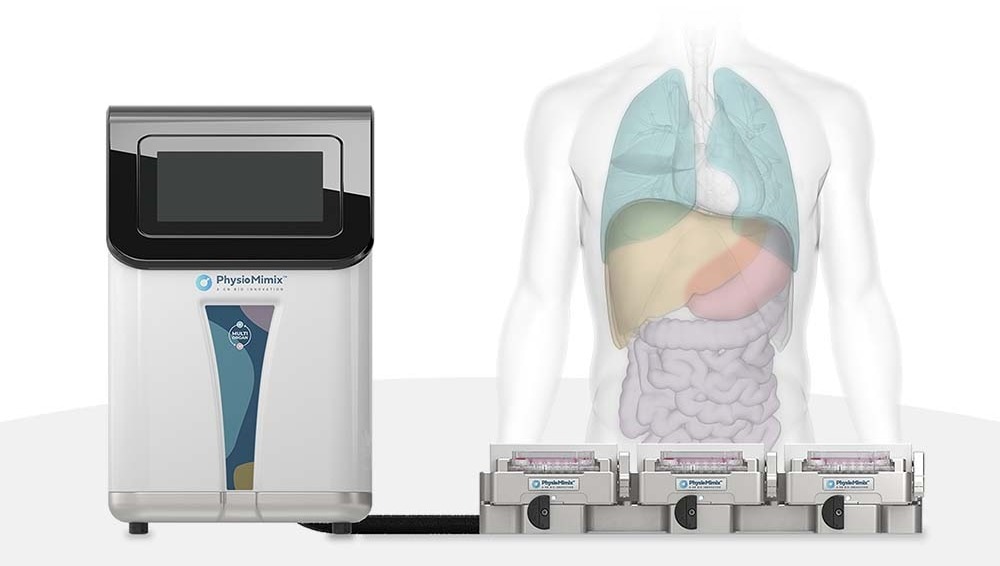
Image Credit: CN Bio Innovations Limited
Microphysiological systems (MPS) are Organ-on-a-chip platforms that use active pumping to provide biologically relevant perfusion throughout the cell culture, which acts as the in vitro equivalent of vascularization. With this media circulation mechanism in place, liver MPS models generate crucial biomechanical information, whilst delivering nutrition and removing waste chemicals from active hepatocytes. These models exhibit improved longevity and functionality compared to other 3D platforms, like spheroids, and have also proven to be more sensitive to hepatotoxic agents.
As discussed above, one of the key factors that dictate the predictability of an imDILI model is the incorporation of a human immune component. It is possible to simulate the innate immune system by including nonparenchymal cells (such as Kupffer cells) when seeding the microtissue. It is, however, difficult to recapitulate the adaptive immune component in static 3D liver models. With a perfused culture system, human peripheral blood mononuclear cells (or other adaptive immune cell types) can be added to the media, which is then circulated throughout the culture, in a similar way to how it would circulate through the body in vivo.
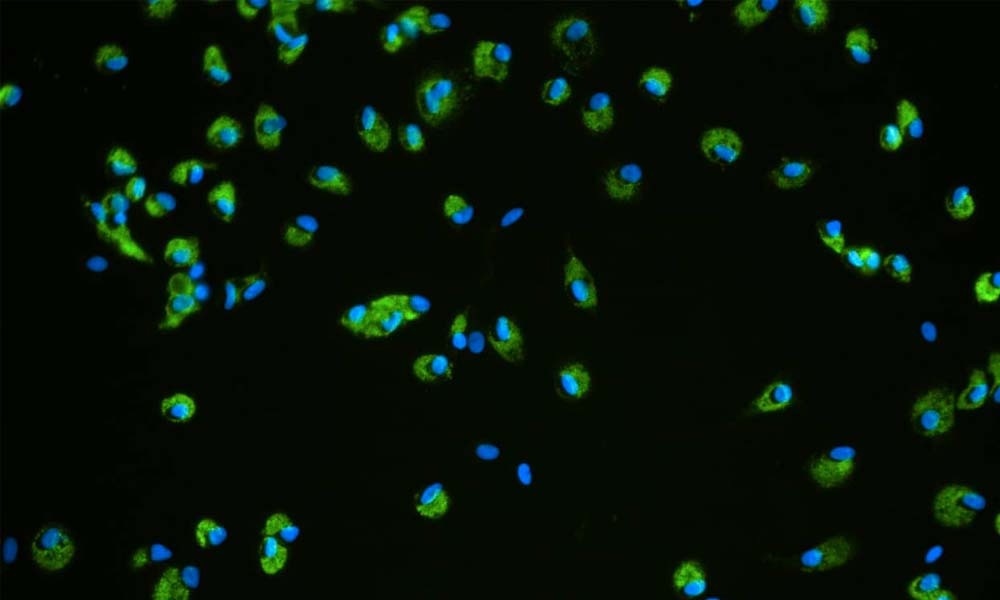
Image Credit: CN Bio Innovations Limited
The addition of the human immune and perfusion components, coupled with easy sample accessibility (inherent to in vitro models), means MPS models are relatively agnostic to drug modality. Bolus dosing of large biologics and oligonucleotides in static culture can often yield a diffusion gradient in the cell’s microenvironment, which is a poor representation of clinical exposure. The development of these MPS models has led us into a new era of drug discovery, allowing us to predict what has historically been considered unpredictable.
As commercial OOC models become more readily available, selecting the right platform for your project becomes increasingly important. The PhysioMimix® range of MPS delivers an open-well solution that enables easy cell monitoring and drug dosing, whilst also being accessible to the average cell culture scientist. Media perfusion and embedded 3D scaffolds ensure hepatocyte functionality and the open-plate architecture of PhysioMimix simplifies the addition of other types of cells, as and when required, to capture adverse drug responses. The PhysioMimix DILI assay has also been extensively characterized and is proven to provide reliable and robust data from both small molecules and new modalities.
About CN Bio
CN Bio is a leading organ-on-a-chip (OOC) company that offers a portfolio of products and contract research services to optimize the accuracy and efficiency of bringing new medicines to market. With more than a decade of research and development experience, they aim to transform the way human-relevant pre-clinical data is generated through the development of advanced in vitro human organ models.
CN-Bio's PhysioMimix® Core microphysiological system (MPS) enables researchers to recreate human biology in the lab and is the only microphysiological system with validated performance across single-, multi-organ, and higher throughput configurations. This easy to adopt, adapt and scale technology bridges the gap between traditional cell culture and human studies, to support the development of safer and more efficacious therapeutics, whilst reducing the dependence on animal model usage.
CN Bio’s portfolio of products (MPS, 3D validated cells, consumable plates) and services support researchers that require reliable, data-rich, in vitro studies, to uncover novel mechanistic insights into drug or disease mechanism of action.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.