Attrition in the therapeutic pipeline can often be associated with the lack of translational efficacy from the pre-clinical phase to the clinic. Organoids demonstrate the significant potential to change the game in disease modeling and drug screening since they closely resemble tissue structure and functionality and demonstrate more predictive responses to drugs. However, there are challenges when trying to practically adopt widespread use of organoids, such as assay complexity, reproducibility, and the limited capability to scale up. This somewhat restricts their use as a primary screening method in drug discovery.
To prevent bottlenecks associated with labor-intensive manual protocols, Molecular Devices UK has developed the CellXpress.ai™ Automated Cell Culture System. This powerful workstation automates the entire organoid culture process for extended and complex workflows.
The CellXpress.ai system can offer media exchange, plating, passaging, organoid monitoring, and endpoint assay execution, as well as the ability to run complex image analysis. In this article, results from the automation of several commonly used organoid protocols, including the culture of 3D organoids in matrix domes, are presented.
Throughout this study, healthy intestinal organoids were cultured, passaged, and expanded in Matrigel® domes (24-well). Cultivation of the organoids was executed using automated media exchange and monitored every 24 hours with the help of machine learning-assisted imaging. After 5–6 days, organoids were collected automatically, purified from Matrigel before being dispersed, then mixed with fresh Matrigel and re-plated.
Subsequently, the organoids were able to self-organize and develop complex crypt structures. To determine organoid number, size (by area), and optical density, organoids were observed in transmitted light and evaluated using machine learning-based image analysis. For endpoint assay (96-well), organoids were stained for viability markers and checked for concentration and time-dependent effects of compounds on healthy intestinal organoids (toxicity evaluation), or patient-derived colorectal cancer organoids (drug screening).
Automated cell culture processes supported by imaging and machine learning-controlled decision-making have significant potential to bring 3D biology to another level, facilitating increased throughput and reproducibility for drug discovery and disease modeling applications.
Methods
Instrumentation
Molecular Devices UK has developed the all-new, powerful CellXpress.ai automated cell culture system, making it possible to automate the entire cell culture process with an integrated incubator, liquid handler, and image-based decision-making. This hands-off approach can manage demanding feeding and passaging schedules by monitoring the development of cell cultures with periodic imaging and analysis, and uses machine learning to trigger any passaging, endpoint assay, or troubleshooting steps.
Fluorescent (FL) images were obtained using the ImageXpress® Confocal HT.ai High-Content Imaging System (Molecular Devices) in combination with MetaXpress® High-Content Image Acquisition and Analysis Software. For intestinal organoids, Z-stack images were captured with the 4X or 10X objectives using confocal mode. MetaXpress or IN Carta® Image Analysis Software was applied across all analyses.
Cell culture protocols
Molecular devices UH has developed a protocol for 3D intestinal organoids* that were taken from primary mouse intestinal cells using existing methods (STEMCELL Technologies). Cells were cultured and differentiated in line with the STEMCELL Technologies protocol. IntestiCult™ Organoid Growth Medium (STEMCELL Technologies) was applied for cell culture.
Cells were seeded in 50 % reduced growth-factor Matrigel or Cultrex (Corning) domes in a 24-well plate format. They were subsequently fed every second day with fresh media over a period of 7–10 days. Intestinal organoids were then passaged, dissociated, and re-plated into fresh Matrigel domes.

Figure 1. The CellXpress.ai cell culture system components and functionality. Image Credit: Molecular Devices UK Ltd
Organoid cell culture automation
Organoid culture in Matrigel domes was conducted in accordance with the basic STEMCELL Technologies guidance protocol for mouse intestinal organoids. Organoids were cultivated and passaged in 24-well plate format with single dome 40–50 μl, 50–60 % Matrigel or Cultrex.
Organoid plating was initiated from the organoids suspended in Matrigel which was set into a pre-chilled 96-well deep well block. The suspension was pipetted using four pipette tips and dispensed into a 24-well plate, four tips at a time. Plating into a 96-well format was also tested.
Organoid feeding was performed by removing spent media and adding fresh media, four wells at a time. Organoid imaging/monitoring was conducted in transmitted light using CellXpress.ai automated cell culture system with 2x or 4x magnification. Image analysis was completed using a machine learning-based protocol. The analysis was able to assess organoid number, mean and total area, density, and several other measurements.
Passaging organoids was set for every four days, or as user-directed, or by automatic decision-making determined by one or more selected measurements (e.g. total area or organoid density).
CellXpress.ai Automated Cell Culture System
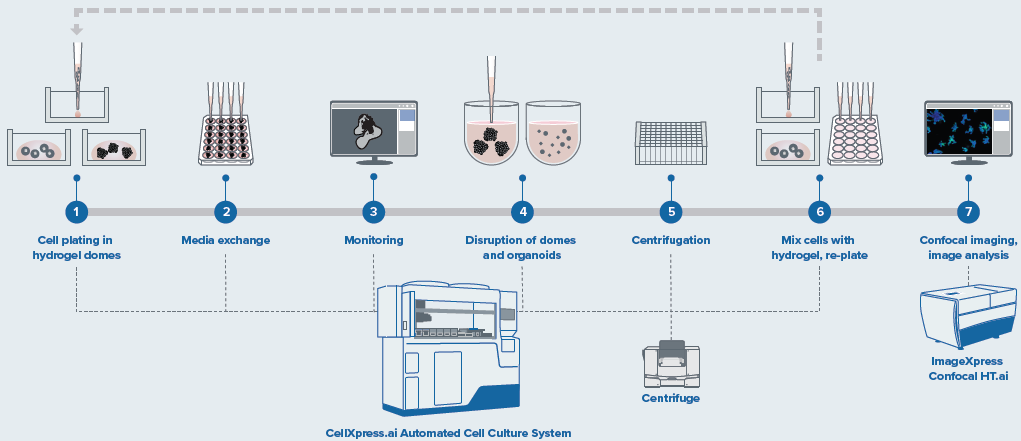
Figure 2. Schematic diagram of automated organoid culture and passaging protocol. Image Credit: Molecular Devices UK Ltd
Organoid passaging was completed using an assortment of pipetting steps and external centrifugation steps to ensure the mouse organoids workflow was optimized. Modification of the flow rates, pipetting steps and repeats, centrifugation speed, etc. can be tailored by changing the necessary “fine-tuning” steps.
For the passaging process, the removal of the media was completed, and Matrigel domes were incubated using Gentle cell dissociation reagent, then the Matrigel domes were broken using rigorous pipetting. The mix was harvested into a 96-deep-well block. Then optional pooling of two wells into one was carried out, followed by centrifugation on an external centrifuge with a speed of 400 g for five minutes.
The block was then repositioned, the media removed, and the organoids were washed once. After a second centrifugation, the majority of supernatant was aspirated, and then, using smaller tips, the organoids were broken with rigorous pipetting. Subsequently, fresh Matrigel was introduced to the appropriate volume, mixed, and seeded into a new plate. Organoid staining/imaging for endpoint assay was performed using FL imaging with the ImageXpress Confocal HT.ai instrument.
Automated organoid workflow
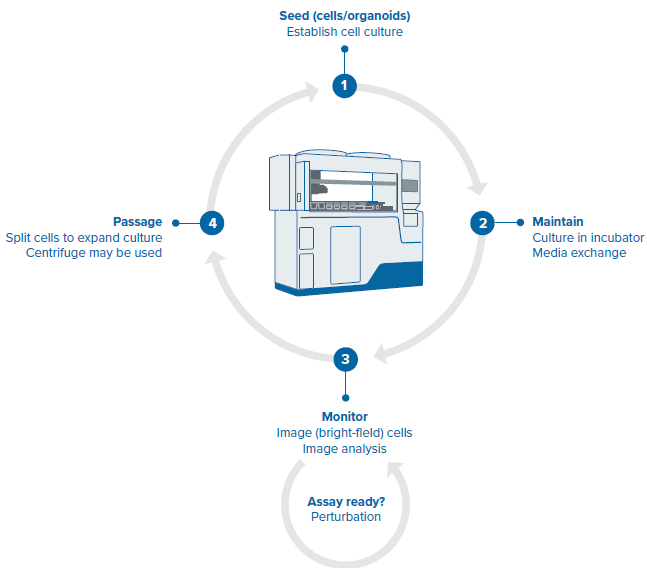
Figure 3. Steps of the organoid culture and passaging protocol. Image Credit: Molecular Devices UK Ltd
- Seeding
Image Credit: Molecular Devices UK Ltd
Matrigel domes are automatically positioned and the ECM suspension is combined with organoids (cells). The seeded domes are configured in 24 well plates, incubated before being transferred to the incubator and moved back to the deck to introduce other media.
- Media exchange
Image Credit: Molecular Devices UK Ltd
Remove media and introduce fresh media. Set the process protocol to repeat the routine every ‘X’ hours.
- Monitoring
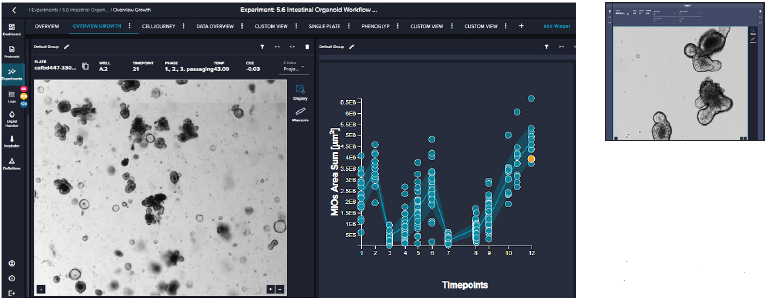
Image Credit: Molecular Devices UK Ltd
The user defines routine imaging of the plate protocols. Real-time image analysis is powered by a pre-trained deep-learning-based model (or customized model). Data analytics tools enable the view of organoids' growth over time. Results can be used to activate other processes/protocols.
- Passaging
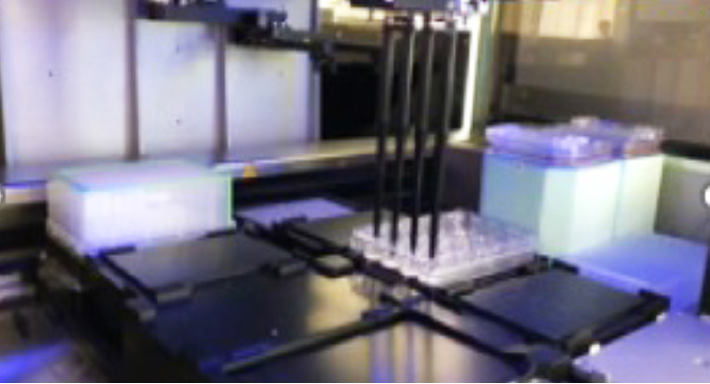
Image Credit: Molecular Devices UK Ltd
Remove media and release organoids from the ECM. Centrifugation runs externally for fragmentation of the organoids. Introduce new ECM/Matrigel and seed into new well plate.
The workflow steps that include seeding, media exchange, monitoring and passaging of intestinal organoids are scheduled with the pre-set “phases” of the protocol. The first phase is seeding, which programs the step of placing Matrigel organoids into 24 well plates. The next phase of the protocol includes feeding/monitoring/passaging steps with user-defined timing and parameters.
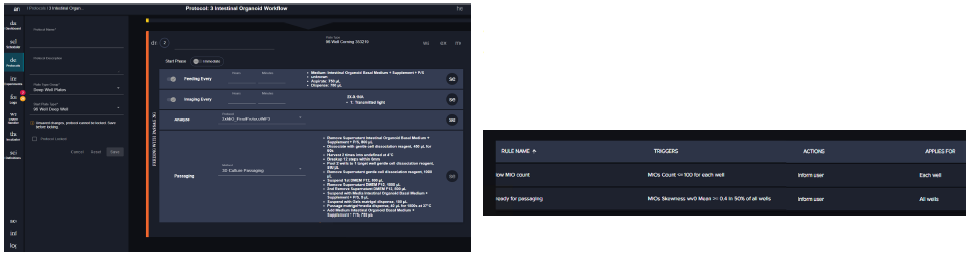
Figure 4. Steps of the organoid culture and passaging software protocol. Image Credit: Molecular Devices UK Ltd
Results
In this study, continuous culture of intestinal organoids was carried out for more than a month. Automated seeding of organoid domes enabled consistent size and precise centering of domes in 24-well plates, which made imaging and image analysis easier and more accurate. Organoids grow as expected throughout the automated culture process, creating protrusions in keeping with typical intestinal organoids morphology.
Across all wells, the distribution and number of organoids remained consistent. Images were captured daily using 2x objective, and image analysis was performed using the pre-trained machine-learning model. The image analysis makes it possible to find organoids and establish the number of organoids, organoid density, roundness, granularity, and other morphological criteria, including averages and total area.
The software makes it easy to review organoid domes throughout the culture process and offers real-time analysis and time course plots representing various measurements. Data can also be exported into an Excel datasheet for further analysis. Figure 6 exhibits changes in organoids' area sum and number of organoids over time.
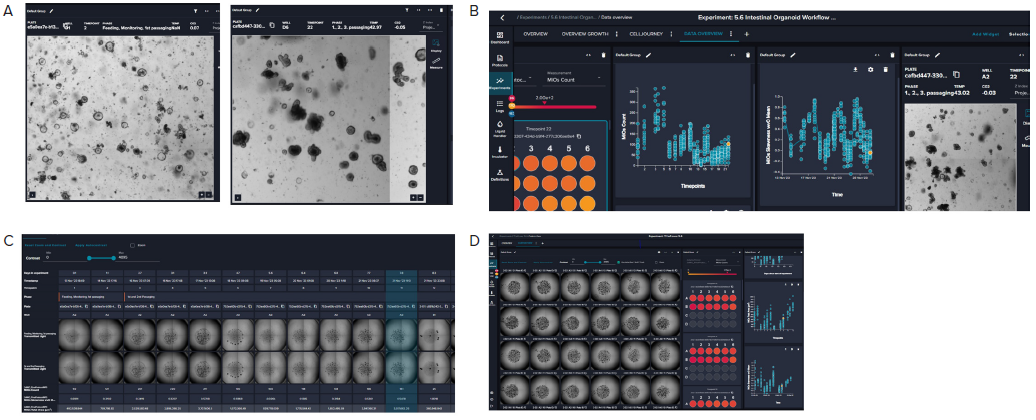
Figure 5. A. Representative images (4X) of organoid culture taken at different time points of the continuous culture. B. Graphical representation
of organoid analysis: organoid count and skewness over time. Skewness includes a combination of optical density, granularity, and other optical
parameters. The value is useful in defining the time for organoid passaging. C. Imaging of a single well over time showing the “cell journey”. D. Tiled 4X
images of organoid domes from a 24-well plate. Image Credit: Molecular Devices UK Ltd
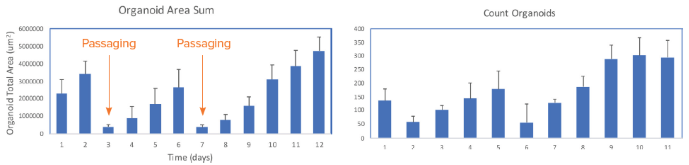
Figure 6. Presentation of organoid count and organoid total area (area sum) over time. Averages and STDEV were calculated from 24 wells of the plate. Image Credit. Molecular Devices UK Ltd
Summary
Organoid technologies have game-changing abilities in disease modeling and drug screening, since they closer resemble tissue structure and functionality, and display more predictive responses to drugs.
The challenges that may emerge with the practical widespread adoption of organoids, such as assay complexity, lack of reproducibility, and the capability to scale up screening have somewhat restricted their rollout as a primary screening method in drug discovery.
However, this study demonstrates how researchers can prevent certain bottlenecks that come with labor-intensive, complex protocols to increase productivity for drug screening.
A new and powerful solution, CellXpress.ai Automated Cell Culture System, give labs the power to automate the entire cell culture process—from assay assembly to screening and data analysis—with machine learning-powered workflows that deliver more reliable and reproducible assays.
Automated cell culture processes powered by imaging and machine learning-controlled decision-making show significant potential in taking 3D biology to the next level by boosting throughput and reproducibility, and allowing for a variety of high-throughput drug discovery and precision medicine applications.
About Molecular Devices UK Ltd
Molecular Devices is one of the world’s leading providers of high-performance life science technology. We make advanced scientific discovery possible for academia, pharma, and biotech customers with platforms for high-throughput screening, genomic and cellular analysis, colony selection and microplate detection. From cancer to COVID-19, we've contributed to scientific breakthroughs described in over 230,000 peer-reviewed publications.
Over 160,000 of our innovative solutions are incorporated into laboratories worldwide, enabling scientists to improve productivity and effectiveness – ultimately accelerating research and the development of new therapeutics. Molecular Devices is headquartered in Silicon Valley, Calif., with best-in-class teams around the globe. Over 1,000 associates are guided by our diverse leadership team and female president that prioritize a culture of collaboration, engagement, diversity, and inclusion.
To learn more about how Molecular Devices helps fast-track scientific discovery, visit www.moleculardevices.com.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.