Parallel temperature initial rates (PTIR) that stem from the chromatographic separation of aggregating protein solutions are merged with continuous simultaneous multiple sample light scattering (SMSLS) to quantitatively deduce the kinetics and underlying mechanisms of protein aggregation.
Image Credit: ustas7777777/Shutterstock.com
PTIR calculates the rates at which originally monomeric proteins are transformed into aggregates at various temperatures under initial-rate settings. Utilizing SMSLS for identical conditions generates time courses of the absolute Rayleigh scattering ratio, IR(t), from which it is possible to calculate a potentially different assessment of aggregation rates.
This article compares such measures of aggregation rates over many solution conditions that lead to various aggregation mechanisms for anti-streptavidin (AS) immunoglobulin gamma-1 (IgG1). The findings demonstrate how both approaches produce complementary data when inferring aggregation mechanisms alongside situations where they deliver novel mechanistic information that was impossible to deduce previously.
Criteria are demonstrated for when both approaches are anticipated to deliver similar outcomes for quantitative rates, the possible limitations when solution non-idealities are large, and a comparison of the temperature dependence of AS-IgG1 aggregation rates with data already published for other antibodies.
Introduction
Proteins are fundamentally unstable molecules that sustain a dynamic equilibrium among folded, unfolded, and partly unfolded states in solution, with just a minute free energy bias for folded states under native-favoring or folded-state favoring settings.1 Proteins including monoclonal antibodies (MAbs) are among the fastest-growing groups of novel drug candidates in the pharmaceutical industry.2
Assessing and optimizing in vitro stability for multiple degradation routes is important in developing therapeutic protein drug products.3
One of the most common routes is known as non-native aggregation, which typically relates to processes through which an otherwise natively folded, monomeric protein can become a part of aggregates composed of partially or completely unfolded protein chains.4
Frequently, the aggregates are made stable by powerful non-covalent contacts that occur between hydrophobic amino acids, alongside hydrogen bonding between the amide backbones of neighboring proteins. Due to this, these non-native aggregates frequently have a high degree of inter-protein beta-sheet structure and are practically irreversible under the solution conditions they form.5,6
Although some early stages of non-native aggregation (referred to as aggregation from here) are commonly reversible, the net aggregation process is not reversible. This necessitates the consideration of aggregation rates and mechanisms or pathways when designing rational approaches to control and enhance the stability of proteins.3,7
There are too many possible aggregation mechanisms to properly summarize in this article. However, work until now suggests that many therapeutic proteins, including MAbs,8-15 antibody fragments,16,17, and cytokines,18,19 have a relatively widespread set of possible aggregation pathways in common that are also used by non-therapeutic proteins.20-22
Figure 1 outlines these for the example of an antibody and is adapted from Refs.6,23
In brief, it is possible for monomeric proteins to partially or completely unfold to uncover hydrophobic sequences that can generate robust inter-protein contacts that stabilize aggregates, also known as aggregation-prone “hot spots”.24,25 This unfolding procedure is reversible if the monomers can refold before coming into contact with a different protein. In most conditions of practical interest, the temperature is far below the midpoint unfolding temperature (Tm) that the unfolding transition(s) will stabilize more quickly than the time scales for successive aggregation events.7 In this situation, the fraction of the monomer population that consists of the partially unfolded or “reactive” (R) state is calculated by the thermodynamics of unfolding. Thus, the net aggregation rate can change by orders of magnitude with a temperature difference of just 5-10 oC. This is because the equilibrium constant for unfolding (K) has a large associated unfolding enthalpy (ΔHun).26,27
Association of R monomers might involve reversible steps before nucleation of the smallest species that are practically irreversible, defined as nuclei and represented by Ax (x = 1⁄4 nucleus stoichiometry) in Figure. 1. Traditionally, a considerable number of the protein and peptide systems that were assessed demonstrated rapid “downhill” polymerization of these originally small aggregates. This made way for the term “nuclei”, by analogy with nucleation and growth in phase transitions.7
More recently, research has shown that proteins, including MAbs, exhibit a broader range of behaviors. Sometimes, they form irreversible dimers (x = 2) but those dimers increase in size at a very slow pace, if at all.8,11,12,28 In the nomenclature of ref.,11,12, this was defined as nucleation-dominated (ND) aggregation and corresponds to practically stopping at stage (3) in Figure 1.
In other cases, nucleation is merged with considerable consumption of monomer through growth by monomer-addition or “chain polymerization” (CP), which corresponds to the merging of stages (1) to (4) in Figure 1.10-12,29 Lastly, when aggregates associate rapidly with one another, it is possible to observe growth through association polymerization (AP) to generate extremely large soluble species, alongside what is practically phase separation (PS) of the aggregates.8,10-12,29-32
In recent times, the effects of solution conditions (pH, NaCl concentration, and buffer species) were systematically calculated for the aggregation mechanisms of an immunoglobulin gamma-1 (IgG1) antibody that targets streptavidin,11,12 and that acts as a test system in this article. For instance, the mechanism alternated between ND, CP, and PS by altering the pH of 4 and 6 with a 5 mM sodium citrate buffer.11,12 That work used ex vivo laser scattering with size exclusion chromatography (SEC) for a single temperature for each solution condition and required several samples at each temperature.
Considerable user manipulation was needed alongside user time and sample material. However, the problem of whether the underlying mechanisms change due to temperature remained unresolved. The current article homes in on a method to circumvent such limitations by merging two newly developed approaches to attain temperature-dependent assessments of aggregation rates: parallel-temperature initial rates (PTIR) with SEC,33 alongside simultaneous multiple-sample static light scattering (SMSLS).34
Parallel temperature initial rates (PTIR) analysis utilizes the following method for calculating degradation rates stemming from temperature; in this situation, the degradation route is aggregation. For context, in conventional methods, one calculates monomer loss for multiple samples at predefined incubation times for a single or small number of temperatures. In the PTIR approach, one calculates monomer loss for a single or small number of samples at multiple temperatures for an identical incubation time.
Elsewhere it has been shown that in the initial rate regime, the two methods are practically the same, although the PTIR approach is more sample sparing and has higher efficiency.33 As aggregation is calculated for the initial rates regime alone, no particular aggregation model is required to be assumed, however they can be deduced later on (see Discussion section). The ultimate aggregation rate coefficient (kobs), or reduced initial aggregation rate (units of 1/time), is typically produced using the following equation:
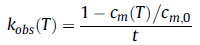 |
(1) |
and is founded on the typical outcome that multiple mechanisms reduce to zero-order kinetics in the limit of small extents of reaction.33,35 In the relations described above, t is the preselected incubation time, T is the incubation temperature, cm is the monomer concentration at T for that incubation time and cm,0 is the initial concentration before heating. In previously conducted work, the ratio of cm to cm,0 was characterized as the monomer fraction (m).12,33
The development of simultaneous multiple static light scattering (SMSLS) was founded on the idea that the multiple approaches can only track a few time points stemming from the need for ex-situ analysis, or they monitor a single sample in-situ without taking advantage of the ability to reconfigure the tool to track several samples simultaneously. SMSLS tracks multiple samples simultaneously and makes it possible to use a related parallel method to what PTIR is based upon. In the limit of low protein concentration (more rigorously, the limit of negligible non-idealities from protein-protein and protein solvent/cosolute interactions), light scattering (LS) produces the weight- averaged molecular weight (Mw) for a sample as required.34,36 Additionally, the weight-averaged molecular weight must produce a divergent “extent of reaction” in comparison to the number-averaged molecular weight- the latter is quantitatively identical to the degree of monomer loss in the limit wherein all aggregates are soluble.22,37
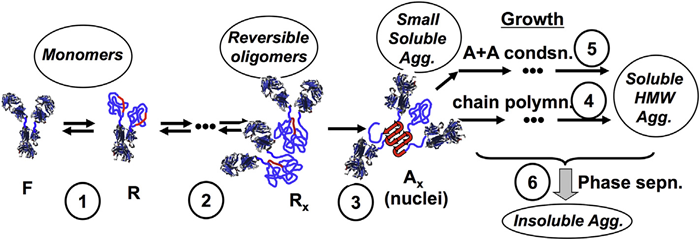
Figure 1. Schematic depiction of multiple stages in non-native aggregation of monoclonal antibodies in solution: (1) partial unfolding to reveal aggregation-prone “hot spots” (red); (2) weak, reversible self-association, prior to (3) structural rearrangement to form effectively irreversible nuclei that then have the potential to grow via (4) monomer addition (termed chain polymerization) or (5) aggregate-aggregate coalescence (association polymerization) and ultimately (6) aggregate phase separation. Adapted from Ref. [6,23].
In many biotechnology industries, quantifying aggregation rates based on the standard of monomer loss (percent mass conversion) is critical.23,38 It is, therefore, interesting from a practical point of view to identify settings where aggregation rates from SMSLS and PTIR can deliver similar outcomes. Fundamentally, it may be helpful to determine criteria and conditions wherein approaches like SMSLS and PTIR can deliver complementary data to track how aggregation mechanisms may be able to change due to temperature and parameters, including solution conditions. The following article serves as a base for future applications of PTIR and SMSLS as a combined technique for calculating protein aggregation rates and identifying aggregation mechanisms spanning a variety of solution conditions and/or temperatures.
What remains in this article is organized as follows: The Materials and Methods section summarizes the experimental methods, data analysis, and a mass-action kinetic model for mechanisms developed beforehand in Figure 1.
Afterward, the Results and Discussion section compares the temperature-dependent rates and time-dependent behaviors for SMSLS and PTIR in the initial-rates regime, which is most significant for practical use cases like analyzing the shelf life of biotechnology products. A comparison between rates from SMSLS and PTIR demonstrates settings wherein the rates quantitatively accord and where they differ significantly, alongside aspects that should be taken into account when comprehending or defining rates from various approaches.
Next comes a discussion of crucial experimental features of various aggregation mechanisms that can be inferred as a function of temperature by merging PTIR and SMSLS alongside conditions where both approaches are complementary versus when they are likely to be useless. The analysis is further demonstrated to provide the threshold oligomer size before phase separation becomes dominant under real-time settings.
This threshold was hypothesized in other theoretical research and indirectly deduced from kinetic modeling of ex-situ aggregation information.10,18,26,39 According to current knowledge, this is the first time it has been directly corroborated in situ in an experimental format.
Lastly, the temperature dependence of aggregation rates for AS-IgG1 is assessed in comparison with those previously cited for various monoclonal antibodies,34 and for the IgG1 Fc domain,17 to demonstrate insights that a quantitative Arrhenius analysis can deliver concerning the unfolding stage(s) of aggregation.
2. Materials and methods
2.1. Materials
Amgen delivered AS-IgG1 at a stock concentration of 30 mg/mL, and 5 mM citrate buffer solutions were produced utilizing MilliQ distilled deionized water (Millipore, Billerica, MA) andcitric acid monohydrate (Fisher Scientific), alongside sodium chloride (Fisher Scientific). The solution pH was modified with sodium hydroxide (Fisher Scientific). Protein solutions were dialyzed against the desired buffer, as earlier discussed.12 Following dialysis, solutions were filtered using 0.22 mm filters (Millipore), and the concentration was calculated by taking into account the absorbance at 280 nm through a UV-vis spectrophotometer (Agilent 8453 UV-Vis, Agilent Technologies, Santa Clara, CA) utilizing the AS-IgG1 extinction coefficient of 1.586 mL/mg cm. All solutions were diluted gravimetrically until final AS-IgG1 concentrations of 1 mg/mL were obtained.
2.2. SMSLS
The SMSLS tool was a prototype from Advanced Polymer Monitoring Technologies, Inc. (New Orleans, LA). It consists of 16 independent sample cells, each carrying its own 660 nm, vertically polarized 30 mW diode laser capable of delivering incident light, alongside independent, programmable temperature control and stepper-motor-controlled stirring, and fiber optic-coupled light scattering detection at 90o. The SMSLS is different from serial measurement tools, as it has no moving optical components, meaning that accurate alignment is permanently maintained for every cell, further allowing the production of absolute assessment of Mw over long periods.34
2.3. PTIR
PTIR was carried out via a tailor-made machine that permits up to 10 independent chambers to be held in parallel at various temperatures, with each having independent Peltier control. Protein solutions were incubated in hermetically sealed HPLC vials (Waters, Milford, MA). Following a certain period, vials were taken out from the PTIR device and quenched on ice to stop the process of aggregation. The remaining monomer (cm) concentration and the initial monomer concentration (cm,0) for each protein solution was calculated utilizing size exclusion chromatography, as discussed above.12,33
The aggregation initial-rate coefficient (kobs) was quantified using Eq. (1), after ensuring that the fractional monomer loss (1 - cm/cm,0) was smaller than 1. The resultant kobs values were quantified as a function of pH, NaCl concentration, and T. They were published in Ref.33.The rates rely heavily on solution conditions (pH and ionic strength) and temperature. Diverse solution conditions are required to change the predominating mechanism. Thus, for aggregation to happen in practical time scales of hours to days, it was important to utilize somewhat diverse temperature ranges for the various solution conditions that make way for the various mechanisms.
References
- C.M. Dobson, Principles of protein folding, misfolding and aggregation, Semin. Cell Dev. Biol. 15 (2004) 3e16, http://dx.doi.org/10.1016/j.semcdb.2003.12.008.
- J.M. Reichert, Antibodies to watch in 2016, mAbs 0 (2015) 1e8, http://dx.doi.org/10.1080/19420862.2015.1125583.
- W. Wang, S. Singh, D.L. Zeng, K. King, S. Nema, Antibody structure, instability, and formulation, J. Pharm. Sci. 96 (2007) 1e26, http://dx.doi.org/10.1002/jps.20727.
- W. Wang, C.J. Roberts (Eds.), Aggregation of Therapeutic Proteins, John Wiley & Sons, Inc., 2010.
- A.L. Fink, Protein aggregation: folding aggregates, inclusion bodies and amyloid, Fold. Des. 3 (1998) R9eR23.
- C.J. Roberts, Therapeutic protein aggregation: mechanisms, design, and control, Trends Biotechnol. 32 (2014) 372e380, http://dx.doi.org/10.1016/j.tibtech.2014.05.005.
- C.J. Roberts, Non-native protein aggregation kinetics, Biotechnol. Bioeng. 98 (2007) 927e938, http://dx.doi.org/10.1002/bit.21627.
- P. Arosio, S. Rima, M. Morbidelli, Aggregation mechanism of an IgG2 and two IgG1 monoclonal antibodies at low pH: from oligomers to larger aggregates, Pharm. Res. 30 (2013) 641e654, http://dx.doi.org/10.1007/s11095-012-0885-3.
- H. Bajaj, V.K. Sharma, A. Badkar, D. Zeng, S. Nema, D.S. Kalonia, Protein structural conformation and not second virial coefficient relates to long-term irreversible aggregation of a monoclonal antibody and ovalbumin in solution, Pharm. Res. 23 (2006) 1382e1394, http://dx.doi.org/10.1007/s11095-006-0018-y.
- R.K. Brummitt, D.P. Nesta, L. Chang, A.M. Kroetsch, C.J. Roberts, Nonnative aggregation of an IgG1 antibody in acidic conditions, part 2: nucleation and growth kinetics with competing growth mechanisms, J. Pharm. Sci. 100 (2011) 2104e2119, http://dx.doi.org/10.1002/jps.22447.
- N. Kim, R.L. Remmele Jr., D. Liu, V.I. Razinkov, E.J. Fernandez, C.J. Roberts, Aggregation of anti-streptavidin immunoglobulin gamma-1 involves Fab unfolding and competing growth pathways mediated by pH and salt concentration, Biophys. Chem. 172 (2013) 26e36, http://dx.doi.org/10.1016/j.bpc.2012.12.004.
- G.V. Barnett, V.I. Razinkov, B.A. Kerwin, T.M. Laue, A.H. Woodka, P.D. Butler, T. Perevozchikova, C.J. Roberts, Specific-ion effects on the aggregation mechanisms and proteineprotein interactions for anti-streptavidin immunoglobulin Gamma-1, J. Phys. Chem. B 119 (2015) 5793e5804, http://dx.doi.org/10.1021/acs.jpcb.5b01881.
- D.D. Banks, R.F. Latypov, R.R. Ketchem, J. Woodard, J.L. Scavezze, C.C. Siska, V.I. Razinkov, Native-state solubility and transfer free energy as predictive tools for selecting excipients to include in protein formulation development studies, J. Pharm. Sci. 101 (2012) 2720e2732, http://dx.doi.org/10.1002/jps.23219.
- S.B. Hari, H. Lau, V.I. Razinkov, S. Chen, R.F. Latypov, Acid-Induced aggregation of human monoclonal IgG1 and IgG2: molecular mechanism and the effect of solution composition, Biochem. (Mosc.) 49 (2010) 9328e9338, http://dx.doi.org/10.1021/bi100841u.
- C.B. Andersen, M. Manno, C. Rischel, M. Thorolfsson, V. Martorana, Aggregation of a multidomain protein: a coagulation mechanism governs aggregation of a model IgG1 antibody under weak thermal stress, Protein Sci. 19 (2010) 279e290, http://dx.doi.org/10.1002/pro.309.
- R.F. Latypov, S. Hogan, H. Lau, H. Gadgil, D. Liu, Elucidation of acid-induced unfolding and aggregation of human immunoglobulin IgG1 and IgG2 Fc, J. Biol. Chem. 287 (2011) 1381e1396, http://dx.doi.org/10.1074/jbc.M111.297697.
- H. Wu, K. Truncali, J. Ritchie, R. Kroe-Barrett, S. Singh, A.S. Robinson, C.J. Roberts, Weak protein interactions and pH- and temperature-dependent aggregation of human Fc1, mAbs 0 (2015) 1e12, http://dx.doi.org/10.1080/19420862.2015.1079678.
- C.J. Roberts, R.T. Darrington, M.B. Whitley, Irreversible aggregation of recombinant bovine granulocyte-colony stimulating factor (bG-CSF) and implications for predicting protein shelf life, J. Pharm. Sci. 92 (2003) 1095e1111, http://dx.doi.org/10.1002/jps.10377.
- R. Thirumangalathu, S. Krishnan, D.N. Brems, T.W. Randolph, J.F. Carpenter, Effects of pH, temperature, and sucrose on benzyl alcohol-induced aggregation of recombinant human granulocyte colony stimulating factor, J. Pharm. Sci. 95 (2006) 1480e1497, http://dx.doi.org/10.1002/jps.20619.
- M. Rosa, C. Lopes, E.P. Melo, S.K. Singh, V. Geraldes, M.A. Rodrigues, Measuring and modeling hemoglobin aggregation below the freezing temperature, J. Phys. Chem. B 117 (2013) 8939e8946, http://dx.doi.org/10.1021/jp4035369.
- M.S. Kosinski-Collins, J. King, In vitro unfolding, refolding, and polymerization of human gammaD crystallin, a protein involved in cataract formation, Protein Sci. 12 (2003) 480e490.
- Y. Li, B.A. Ogunnaike, C.J. Roberts, Multi-variate approach to global protein aggregation behavior and kinetics: effects of pH, NaCl, and temperature for achymotrypsinogen A, J. Pharm. Sci. 99 (2010) 645e662, http://dx.doi.org/10.1002/jps.21869.
- C.J. Roberts, T.K. Das, E. Sahin, Predicting solution aggregation rates for therapeutic proteins: approaches and challenges, Int. J. Pharm. 418 (2011) 318e333, http://dx.doi.org/10.1016/j.ijpharm.2011.03.064.
- A. Caflisch, Computational models for the prediction of polypeptide aggregation propensity, Curr. Opin. Chem. Biol. 10 (2006) 437e444, http://dx.doi.org/10.1016/j.cbpa.2006.07.009.
- N.J. Agrawal, S. Kumar, X. Wang, B. Helk, S.K. Singh, B.L. Trout, Aggregation in protein-based biotherapeutics: computational studies and tools to identify aggregation-prone regions, J. Pharm. Sci. 100 (2011) 5081e5095, http://dx.doi.org/10.1002/jps.22705.
- C.J. Roberts, Kinetics of irreversible protein aggregation: analysis of extended lumry-eyring models and implications for predicting protein shelf life, J. Phys. Chem. B 107 (2003) 1194e1207, http://dx.doi.org/10.1021/jp026827s.
- A. Saluja, V. Sadineni, A. Mungikar, V. Nashine, A. Kroetsch, C. Dahlheim, V.M. Rao, Significance of unfolding thermodynamics for predicting aggregation kinetics: a case study on high concentration solutions of a multi-domain protein, Pharm. Res. (2014), http://dx.doi.org/10.1007/s11095-013-1263-5.
- P. Arosio, B. Jaquet, H. Wu, M. Morbidelli, On the role of salt type and concentration on the stability behavior of a monoclonal antibody solution, Biophys. Chem. 168e169 (2012) 19e27, http://dx.doi.org/10.1016/j.bpc.2012.05.004.
- E. Sahin, A.O. Grillo, M.D. Perkins, C.J. Roberts, Comparative effects of pH and ionic strength on protein-protein interactions, unfolding, and aggregation for IgG1 antibodies, J. Pharm. Sci. 99 (2010) 4830e4848, http://dx.doi.org/10.1002/jps.22198.
- P. Arosio, S. Rima, M. Lattuada, M. Morbidelli, Population balance modeling of antibodies aggregation kinetics, J. Phys. Chem. B 116 (2012) 7066e7075, http://dx.doi.org/10.1021/jp301091n.
- E. Sahin, W.F. Weiss, A.M. Kroetsch, K.R. King, R.K. Kessler, T.K. Das, C.J. Roberts, Aggregation and pH-temperature phase behavior for aggregates of an IgG2 antibody, J. Pharm. Sci. (2012), https://jpharmsci.org/article/S0022-3549(15)31598-7/abstract.
- R.K. Brummitt, D.P. Nesta, L. Chang, S.F. Chase, T.M. Laue, C.J. Roberts, Nonnative aggregation of an IgG1 antibody in acidic conditions: part 1. Unfolding, colloidal interactions, and formation of high-molecular-weight aggregates, J. Pharm. Sci. 100 (2011) 2087e2103, http://dx.doi.org/10.1002/jps.22448.
- G.V. Barnett, V.I. Razinkov, B.A. Kerwin, A. Hillsley, C.J. Roberts, Acetate- and citrate-specific ion effects on unfolding and temperature-dependent aggregation rates of anti-streptavidin IgG1, J. Pharm. Sci. 105 (2016) 1066e1073.
- M.F. Drenski, M.L. Brader, R.W. Alston, W.F. Reed, Monitoring protein aggregation kinetics with simultaneous multiple sample light scattering, Anal. Biochem. 437 (2013) 185e197, http://dx.doi.org/10.1016/j.ab.2013.02.014.
- K.J. Laidler, Chemical Kinetics, second ed., McGraw, 1965.
- W.H. Stockmayer, Light scattering in multicomponent systems, J. Chem. Phys.18 (1950) 58e61, http://dx.doi.org/10.1063/1.1747457.
- E. Sahin, C.J. Roberts, Size-exclusion chromatography with multi-angle light scattering for elucidating protein aggregation mechanisms, Methods Mol. Biol. 899 (2012) 403e423.
- V. Kayser, N. Chennamsetty, V. Voynov, B. Helk, K. Forrer, B.L. Trout, Evaluation of a non-arrhenius model for therapeutic monoclonal antibody aggregation, J. Pharm. Sci. 100 (2011) 2526e2542, http://dx.doi.org/10.1002/jps.22493.
- I. Arora, R. Bansal, V. Joshi, A.S. Rathore, Aggregation kinetics for monoclonal antibody products, Inter J. Chem. Eng. Appl. 5 (2014) 433e438.
- J.M. Andrews, C.J. Roberts, Non-native aggregation of a-chymotrypsinogen occurs through nucleation and growth with competing nucleus sizes and negative activation energies, Biochem. (Mosc.) 46 (2007) 7558e7571, http://dx.doi.org/10.1021/bi700296f.
- W.F. Weiss IV, T.K. Hodgdon, E.W. Kaler, A.M. Lenhoff, C.J. Roberts, Nonnative protein polymers: structure, morphology, and relation to nucleation and growth, Biophys. J. 93 (2007) 4392e4403, http://dx.doi.org/10.1529/biophysj.107.112102.
- J.M. Andrews, C.J. Roberts, A Lumry-Eyring nucleated polymerization model of protein aggregation kinetics: 1. Aggregation with pre-equilibrated unfolding, J. Phys. Chem. B 111 (2007) 7897e7913.
- Y. Li, C.J. Roberts, Lumry�Eyring nucleated-polymerization model of protein aggregation kinetics. 2. Competing growth via condensation and chain polymerization, J. Phys. Chem. B 113 (2009) 7020e7032, http://dx.doi.org/10.1021/jp8083088.
- H. Wu, H. Kroe-Barrett, S. Singh, A.S. Robinson, C.J. Roberts, Competing aggregation pathways for monoclonal antibodies, FEBS Lett. 588 (2014) 936e941.
- S. Amin, G.V. Barnett, J.A. Pathak, C.J. Roberts, P.S. Sarangapani, Protein aggregation particle formation, characterization & rheology, Curr. Opin. Colloid Interface Sci. 19 (2014) 438e449, http://dx.doi.org/10.1016/j.cocis.2014.10.002.
- V.N. Uversky, A.L. Fink, Conformational constraints for amyloid fibrillation: the importance of being unfolded, Biochim. Biophys. Acta Proteins Proteomics 1698 (2004) 131e153, http://dx.doi.org/10.1016/j.bbapap.2003.12.008.
- W.F. Weiss 4th, T.M. Young, C.J. Roberts, Principles, approaches, and challenges for predicting protein aggregation rates and shelf life, J. Pharm. Sci. 98 (2009) 1246e1277, http://dx.doi.org/10.1002/jps.21521.
- M.A. Blanco, T. Perevozchikova, V. Martorana, M. Manno, C.J. Roberts, Proteinprotein interactions in dilute to concentrated solutions: a-chymotrypsinogen in acidic conditions, J. Phys. Chem. B 118 (2014) 5817e5831, http://dx.doi.org/10.1021/jp412301h.
- F. Ferrone, Analysis of protein aggregation kinetics, Methods Enzymol. 309 (1999) 256e274.
About Yokogawa Fluence Analytics
Yokogawa Fluence Analytics, which was named as a Top 50 global advanced manufacturing startup by CB Insights, provides patented process analytics and control solutions to polymer and biopharmaceutical customers worldwide. Yokogawa Electric Corporation acquired Fluence Analytics in January 2023.
Yokogawa Fluence Analytics is a global leader in real-time polymer reaction monitoring and control, and its industry-leading ACOMP product is the only commercially available smart manufacturing system that continuously monitors and measures polymerization reactions.
The company’s biopharmaceutical product line includes a high-throughput static light scattering instrument called ARGEN. ARGEN can independently measure the stability of biopolymers under thermal, chemical, and mechanical (physical) stress, while also performing shelf-life stability studies at low temperatures.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.