Quality food and its effect on health are becoming more important to customers. Together with food scandals of the recent past, this pushes the food industry to make their products more transparent.
The origin of raw materials is one key issue. This point touches topics such as the utilization of materials derived from genetically modified organisms and also the use of the correct materials stated on the list of ingredients.
An effective technique that can be used to investigate the genetic background of ingredients and the origin of materials in processed food are technologies based on Polymerase Chain Reaction (PCR), as these techniques are extremely sensitive.
A certain amount and quality of DNA are required to employ PCR-based technologies for the analysis of food. Some sample materials (e.g. plant samples, processed food) contain significant levels of inhibitors, which inhibit PCR-based downstream applications and so interfere with the analysis.
DNA was extracted from northern shrimp, two different kinds of cheese, and hazelnut in the experiments outlined below. DNA was extracted by utilizing innuPREP DNA Kit-IPC16 and innuPREP Food DNA Kit-IPC16.
Extraction was carried out using the InnuPure® C16/C16 touch. Lysed samples were transferred in the Reagent Plastics of the kits after homogenization (optional) and lysis. Reagent Plastics of the kits are pre-filled with all of the required magnetic particles and buffers for DNA extraction.
The Reagent plastics were loaded in the InnuPure® C16/C16 touch followed by fully automated extraction. DNA quality was analyzed after extraction by using gel electrophoresis, spectrophotometry, and quantitative real-time PCR (qRT-PCR) for species analysis.
The automated purification and high pre-processing state of the kits (pre-filled, sealed buffer reservoirs) decreases the hands-on time of the customer. Other than the advantage of reduced time consumption, pre-processing also decreases the risk of human errors and loss of valuable samples.
The analysis of extracted DNA by agarose gel electrophoresis, spectrophotometry, and qRT-PCR analysis shows that DNA extracted with devices and kits from Analytik Jena can be readily employed for relevant downstream applications.
Materials and methods
Using SpeedMill PLUS and Lysis Tubes P (Analytik Jena), the frozen northern shrimps (50 mg) were homogenized in 200 µl H2O for 15 seconds. Homogenates were mixed with 20 µl Proteinase K and 200 µl Lysis Solution CBV and incubated at 50 °C shaking at 600 rpm for 60 minutes.
400 µl of the lysate was transferred to the Reagent Plastic of the innuPREP DNA KitIPC16 before the extraction with InnuPure® C16. 100 mg and 200 mg ground hazelnut were re-suspended in 20 µl Proteinase K and 800 µl Lysis Solution CBV and the samples were lysed at 65 °C for 60 minutes.
400 µl supernatants were used for extraction after lysis using the innuPREP Food DNA KitIPC16 and InnuPure® C16. DNA was extracted from the two types of commercially available cheese. Cheeses were discovered to be derived from cow and goat milk.
Extraction was carried out in quadruplicates by utilizing the InnuPure® C16 touch and innuPREP Food DNA KitIPC16. For each replicate, 200 mg of cheese was employed. The elution volume was set to 100 µl.
Extracts were analyzed by photometry after extraction via the ScanDrop® 250 and qRT-PCR. For qRT-PCR qTOWER3 and innuDETECT Cheese Assay were employed. The analysis was carried out using the Endpoint technique supplied by qPCRsoft.
Samples and reagents
- Lysis Tubes P
- innuDETECT Cheese Assay
- innuPREP DNA Kit-IPC16
- Commercially available kit for hazel analysis
- innuPREP Food DNA Kit-IPC16
Instrumentation
- qTOWER3 G
- InnuPure® C16
- SpeedMill PLUS
- InnuPure® C16 touch
- ScanDrop® 250
- BioShake iQ
- Standard equipment for agarose gel electrophoresis and gel documentation
Results and discussion
DNA quality and integrity were assessed by spectrophotometric analysis after extraction of DNA from frozen shrimp samples. This was performed with the ScanDrop® 250 and agarose gel electrophoresis.
Table 1. Spectrophotometrical analysis of DNA extracted from shrimp samples.
| Sample number |
A260/A280 |
c [ng/µl] |
cmean±SD [ng/µl] |
| 1 |
2.32 |
23.64 |
22.1±1.8 |
| 2 |
1.96 |
21.26 |
| 3 |
2.25 |
23.51 |
| 4 |
1.87 |
19.94 |
In combination with the InnuPure® C16, the innuPREP DNA Kit-IPC16 enables the automated DNA extraction from shrimps with minimal hands-on time. Pure DNA with A260/A280 ratios ranging from 1.87 to 2.32 is gathered as a result of extraction.
For PCR-based downstream applications mean yields of 22.1 ng/µl are sufficient. Agarose gel analysis demonstrates that a part of the DNA is degraded which appears as a smear in lanes 2–5 of Figure 1.
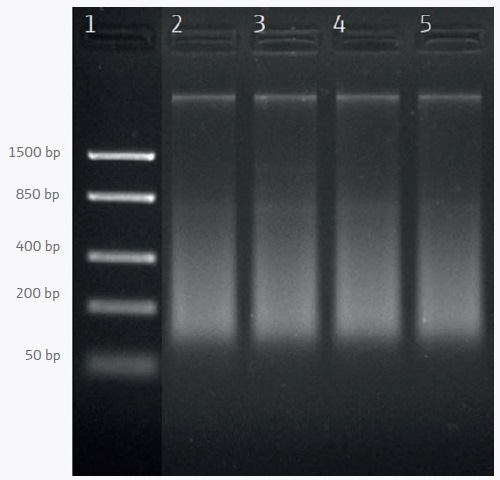
Figure 1. Agarose gel electrophoresis of DNA extracted from frozen shrimp samples. Lane 1: DNA ladder; lanes 2–5: Samples 1–4.
DNA degradation is a natural process that begins shortly after the death of the animals and ends with freezing. Besides degraded DNA, some high molecular weight DNA is visible as a distinct band with a size that is bigger than 1500 bp.
Extracts were employed for qRT-PCR-based detection of hazelnut DNA after the DNA extraction from 100 mg and 200 mg ground hazelnut with the innuPREP Food DNA Kit-IPC16.
Aliquots of the extract were diluted 1:10 followed by amplification of 1 µl undiluted and 1:10 diluted extract in order to detect inhibitory influences of substances contained in the extracted DNA.
DNA extraction from both 100 mg and 200 mg ground hazelnut is possible, as shown in Figures 2 and 3 and it yields DNA that can be used readily for qRT-PCR. With respect to the extraction and contamination of the extracted DNA with PCR inhibitors, sample materials that possess high contents of lipids are often challenging.
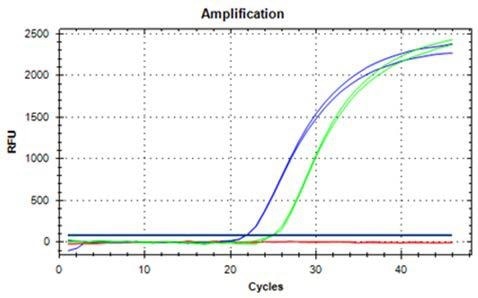
Figure 2. DNA extracted from 100 mg ground hazelnut, amplified, and detected with qRT-PCR in duplicates (blue curves undiluted, green curves 1:10 dilution).
-1-1.jpg)
Figure 3. DNA extracted from 200 mg ground hazelnut, amplified, and detected with qRT-PCR in duplicates (blue curves undiluted, green curves 1:10 dilution).
PCR inhibition can be excluded as a ten-fold dilution of the sample for the extraction of DNA from hazelnut. It leads to a shift of the Ct value of around 3.3 cycles which is the case for extracts from 100 mg and 200 mg, respectively.
DNA was extracted from 200 mg cheese as outlined above. After extraction, extracts were analyzed via photometry by utilizing the ScanDrop® 250 and qRT-PCR. qTOWER3 and innuDETECT Cheese Assay were used for qRT-PCR. Analysis was carried out via the Endpoint technique supplied by qPCRsoft.
Results show that automated extraction via the InnuPure® C16 touch together with the innuPREP Food DNA Kit-IPC16 can be utilized to prepare DNA from cheese. As shown in Figure 4-6, quality and quantity are sufficient for subsequent qRT-PCR application.
-2.jpg)
Figure 4. Detection of a sheep-specific target gene.
-1.jpg)
Figure 5. Detection of a goat-specific target gene.
.jpg)
Figure 6. Detection of a cow-specific target gene.
Analyzing 200 mg starting material leads to eluates which have concentrations between 9-16 ng/µl. Furthermore, using the innuPREP Food DNA Kit-IPC16, proteins are totally removed as indicated by ratios A260/A280 in Table 2.
Table 2. Photometric analysis of nucleic acid extracted from cheese samples.
| Cheese type |
Replicate |
A260/A280 |
c [ng/µl] |
| Goat |
1 |
2.03 |
9.86 |
| 2 |
2.02 |
11.07 |
| 3 |
2.01 |
12.56 |
| 4 |
2.01 |
12.59 |
| Cow |
1 |
2.07 |
16.3 |
| 2 |
1.96 |
12.78 |
| 3 |
2.03 |
12.38 |
| 4 |
2.01 |
9.84 |
The samples derived from milk with known origin were subjected to qRT-PCR analysis of sheep-, goat- and cow-specific genes. Internal positive control (light red) was detected in all instances.
Yet, cow-specific genes were only detected in cheese declared to be derived from cow milk while goat-specific genes were only detected in cheese declared to be derived from goat milk. None of the samples were positive for sheep-specific genes.
Conclusion
In summary, the results show that a combination of InnuPure® C16 touch and DNA extraction kits for this device can be utilized for DNA extraction from a variety of samples related to the food industry.
DNA extracted with this system has the highest quality. Most importantly, DNA extracted with InnuPure C16® touch and the relevant kits can readily be used for qRT-PCR.
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About life science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results.
This will surely ease your daily work and speed up your work processes in a certain way. All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About analytical instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems which dates back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About lab automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness and simplicity.
Moreover, the Automation Team designs, produces and installs fully automated systems tailored to our clients' application, throughput and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems we handle your compounds, biomolecules and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.