Presently, analytical molecular techniques are established tools for the determination of DNA in foods. Identification of accurate and extremely sensitive animal species, and the detection of substitutes are two of the major challenges in the food industry to ensure a high level of food and nutrition quality as well as authenticate unintended ingredients such as pork.
Analytical techniques have to meet specific quality criteria in order to gain confidence and acceptance in the reliability of results obtained by such techniques from various laboratories. The TaqMan®-based innuDETECT Pork Assay allows a highly sensitive analysis of pork species. Further validation studies were conducted to establish extended performance measurement for characterizing the innuDETECT Pork DNA Assay.
Challenge
Reliable detection of porcine DNA in processed and fresh food.
Solution
innuDETECT Pork Assay is used for detecting pork DNA quantitatively and qualitatively.
Materials and Methods
Samples and Reagents
For reference 0.1% (w/w) pork in beef meat standard (Eurofins), genomic DNA of Sus scrofa domestica with a concentration of 100 copies/µl (Eurofins) as well as an in-house developed plasmid with synthetic pork DNA insert (GeneArt) to be used as positive control (available as innuDETECT Animal Quantification Tool (845-IDF-0140100)) were applied with the innuDETECT Pork Assay (845-IDF-0010096).
Instrumentation
The analysis was performed on the real-time PCR platform qTOWER³ from Analytik Jena, on the Step-One Plus from ABI, and on the CFX96 from Bio-Rad to enable performance evaluation for the assay.
Procedure
The 0.1% (w/w) pork in beef meat standard was extracted using the innuPREP DNA Mini Kit (845-KS-1040250) to demonstrate that the developed technique matches with the required performance criteria of detection tests and also to validate the innuDETECT Pork Assay. The protocol is based on the extraction of spin filter nucleic acid along with a patented chemistry based on a certain mixture of non-chaotropic and chaotropic salts (Dual-Chemistry Technology®). Further analysis of the extracted nucleic acids was conducted as stated in instructions for use of the innuDETECT Pork Assay. Through real-time PCR analysis, target amplification and detection were carried out using the HEX channel for internal control and the FAM channel for the target. Table 1 outlines the PCR program.
Table 1. PCR program for qTOWER3
| Step |
Cycle |
Profile |
Temperature |
Holding time |
| 1 |
1 |
Initial denaturation |
95 °C |
120 sec |
| 2 |
35 |
Denaturation |
95 °C |
10 sec |
| Annealing/Elongation * |
45 °C |
45 sec |
* Data acquisition: Fluorescence detection (FAM; HEX)
The analytical sensitivity was determined by analyzing clarified samples with a small quantity of DNA. Extracted nucleic acids from 0.05% pork in beef meat, 10 copies of synthetic plasmid DNA, and one genome equivalent of genomic DNA of Sus scrofa domestica served as templates. A minimum of 30 replicates of each sample type were examined and at least 95% should be detected positive.
The linear range for the innuDETECT Pork Assay was ascertained by analyzing a dilution series of plasmid DNA ranging from 3x100 to 3x107 copies per reaction, and porcine DNA ranging from 3x100 to 3x105 genome equivalents per reaction.
The robustness was tested for the PCR system. Using several PCR instruments, a minimum of 10 samples from 0.1% pork in beef meat were amplified, with deviations in MasterMix compilation and with differing annealing temperatures.
Results and Discussion
Different performance parameters have to be determined in order to use a detection kit for routine analysis. Comparative analysis was carried out for robustness, amplification efficiency as well as the limit of detection. The application of a variety of food samples was also performed.
Detection of Minimal DNA Amounts per Sample
Different starting materials with minimum concentrations of porcine DNA per sample (n=30) were examined for determining the analytical sensitivity of the innuDETECT Pork Assay. Initial results show that 0.05% (w/w) pork in beef meat can be detected. For all replicates (30/30), positive results were determined. Moreover, less than one genome equivalent per reaction could be confirmed using the innuDETECT Pork Assay. Since Pork DNA fragments were demonstrated with amplification of synthetic plasmids, they are adequate for positive test results. As much as 96% could be detected positively using 10 copies of porcine DNA per reaction. All results are outlined in Table 2.
Table 2. Results of analysis of samples with a defined concentration of porcine DNA
| Device |
Pork DNA detection |
Minimal detected amount of DNA |
| Pork in beef meat |
| qTOWER3 |
30/30 |
≤ 0.05% (w/w) pork in beef meat |
| CFX96 |
30/30 |
| ABI Step-One Plus |
30/30 |
| Genomic DNA of Sus scrofa domestica |
| qTOWER3 |
30/30 |
≤ 1 genome equivalent* |
| CFX96 |
30/30 |
| ABI Step-One Plus |
30/30 |
| Synthetic plasmid |
| qTOWER3 |
28/30 |
10 copies |
| CFX96 |
30/30 |
| ABI Step-One Plus |
29/30 |
* Amplification of mitochondrial gene, i.e. several copies per genome
The Detection Range of the innuDETECT Pork Assay
The linear range of the detection assay was ascertained by analyzing synthetic plasmid ranging from 3x100 to 3x107 copies per reaction and porcine DNA ranging from 3x100 to 3x105 genomic equivalents per reaction using the ABI Step-One Plus, the CFX96, and the qTOWER3. The obtained results, as illustrated Figure 1, cover a linear range over 7 log10 steps. On the basis of the Ct value, the slope of the regression line was used for determining the efficiency of the PCR technique.
About 94% efficiency could be detected for qTOWER3 using the positive control plasmid as a template. No criteria of acceptance are defined for qualitative singleplex qPCR techniques. By the measure of a quantitative PCR technique, the efficiency of PCR should range from 90% to 110%. In the meantime, the innuDETECT Pork Assay achieves an optimal result (Broeders et al., 2014). The calculated linearity R2 = 0.9955, an extra criterion for method validation (R2 greater than or equal to 0.98), underlines the assay performance.
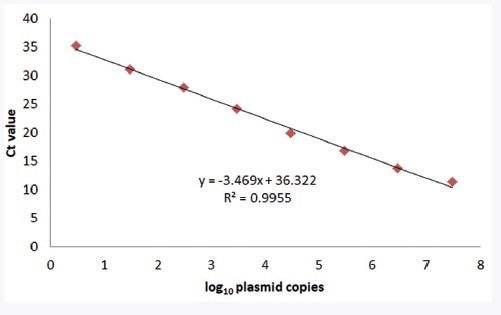
Figure 1. Linearity of the innuDETECT Pork Assay on qTOWER3 using plasmid DNA. The linear regression coefficient (R2) is 0.9955.
Requirement of a Robust qPCR Setup
Different experimental conditions were altered to assess the robustness of the innuDETECT Pork Assay. Therefore, 0.1% pork in beef meat was examined (n=8) using various PCR instruments (CFX96, qTOWER3, ABI Step-One Plus), with changes in MasterMix compilation (+/-30%) as well as with different annealing temperatures (+2 °C/-4 °C).
The modified experimental conditions had no impact on positive assay results. The effects of different annealing temperatures on DNA detection are shown in Figure 2. Deviation in the MasterMix compilation and annealing temperature for all qPCR instruments has an effect on real-time PCR results below 5%. The performance of the innuDETECT Pork Assay is satisfying with regard to these experimental conditions.
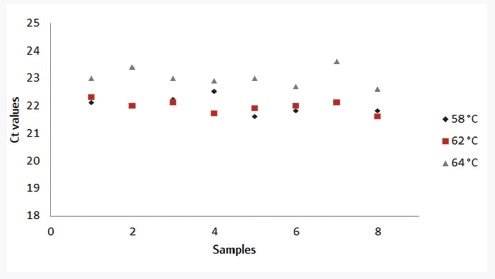
Figure 2. Influence of varying annealing temperature (black: 58 °C, red: 62 °C, grey: 64 °C) on the assay performance, analysis of 0.1% pork in the beef meat (n=8) on qTOWER3 using the innuDETECT Pork Assay.
Analytical Specificity and Application of innuDETECT Pork Assay
The innuDETECT Pork Assay was used to analyze processed and fresh food. For declared mixed, minced meat pork DNA could be detected. The comparative analysis of non-halal and halal marshmallows was also conducted. The results show that the halal-certified marshmallows are free of porcine DNA. In addition to proof of principle, the specificity of the innuDETECT Pork Assay was examined. Thus, kosher beef with and without contamination of pork DNA was used as sample material for verifying the workflow. In the case of the innuDETECT Pork Assay, even the use of contaminated knives for sample preparation is adequate to obtain false positive results.
| Sample |
Result of innuDETECT Pork Assay |
| Minced Meat, mixed |
+ |
| Marshmallow halal |
- |
| Marshmallow non-halal |
+ |
| Kosher beef |
- |
| Kosher beef slices with pork contaminated knife |
+ |
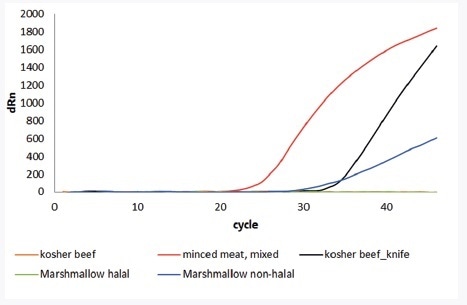
Figure 3. Application of innuDETECT Pork Assay analyzing different fresh and processed food. Top: summarized samples and results; Bottom: Amplification plot; Detection performed on Bio-Rad’s CFX96.
Conclusion
Real-time PCR has been recognized as an accurate, reliable, and rapid application for the identification of animal species. Analytik Jena provides a solution for detecting porcine DNA by using the innuDETECT Pork Assay on qTOWER3 and also on other qPCR instruments. Different performance criteria were analyzed to validate the detection assay. The powerful innuDETECT Pork Assay was convincing enough with a detection limit of ≤ 0.05% (w/w) pork in beef meat. The assay also enabled the effective detection of porcine DNA in processed and fresh food.
References
Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N. and Morisset, D.; GUIDELINES FOR VALIDATION OF QUALITATIVE REAL-TIME PCR METHODS. Trends Food Sci Technol. 2014, 37, pages 115-126.
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results.
This will surely ease your daily work and speed up your work processes in a certain way. All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems which dates back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness and simplicity.
Moreover, the Automation Team designs, produces and installs fully automated systems tailored to our clients' application, throughput and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems we handle your compounds, biomolecules and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.