Automation of library preparation is highly crucial for decreasing the possibilities of error, increasing reproducibility, and minimizing the amount of hands-on time, enabling scientists to produce sequence data from DNA more competently.
By making use of the CyBio® FeliX, a benchtop instrument for a range of automated liquid handling tasks, the HaloPlex Target Enrichment System Kit for NGS library preparation for Ion Torrent™ Sequencing was automated to realize reliable final prepared libraries with high accuracy for downstream sequencing. This article explains the successful proof of principle to produce high-quality DNA libraries on CyBio® FeliX for personal genome analysis with the Ion Torrent™ Sequencing System.
Analytik Jena’s liquid handling series comprises of CyBio® FeliX, a compact and adaptable liquid handling platform for a range of automated liquid handling tasks. The system has 12 deck positions on two levels for tube racks, microplates, reservoirs, and tips on smallest footprint, including accessories such as magnet adapter, thermal shaker, and gripper.
Library preparation is an important, hands-on, and time-taking step in the Next Generation Sequencing (NGS) workflow. To make the NGS library preparation process easy, Analytik Jena designed a protocol for the HaloPlex Target Enrichment System Kit with less user involvement to prepare eight library samples for multiplexed sequencing on the Ion Torrent™ platform.
The CyBio® FeliX liquid handling platform was being tested by the Institute for Laboratory Medicine in the Donauspital in Vienna to prepare DNA sample libraries from patient samples. The efficiency of Next Generation Sequencing is significantly improved by the target enrichment of genomes, enabling scientists to concentrate on particular regions of interest. The Kit is suitable for deep sequencing of small panels of genes (up to 2.5 Mb), for example, those needed for studies of inherited disorders, infectious diseases, cancer, and neurologic diseases.
Challenge
Automated library preparation using the HaloPlex Target Enrichment System Kit for Ion Torrent™ Sequencing.
Solution
The compact and flexible CyBio® FeliX pipetting system was used to carry out all liquid handling steps of the library preparation workflow for personal genome analysis with the Ion Torrent™ Sequencing System.
Materials and Methods
Consumables
- 8-well PCR strip tubes with caps (Nippon Genetics p/n FG-088WF)
- Hard shell 96-well PCR plate (Eppendorf twin.tec, skirted, 0030 128.680)
- Skirted 0.5 mL tubes (STARLAB, E1405-2120)
- 2 mL tubes (STARLAB, E1420-2340)
- Tube Caps (STARLAB, 1480-0199)
- Qubit assay tubes (Life Technologies p/n Q32856)
- CyBio® TipBox 96/50 μL (OL 3811-25-535-N)
- CyBio® TipBox 96/250 μL (OL 3811-25-637-S)
Kits and Reagents
- Library preparation for Ion Torrent™ Sequencing was carried out using eight normalized patient DNA samples (225 ng), isolated using the innuPREP Blood DNA Mini Kit (Analytik Jena AG). Patient DNA was measured using the Qubit fluorometer (Life Technologies). For every to-be-sequenced sample (patient DNA), an individual target-enriched, barcoded library was made ready. The HaloPlex probes were developed using Agilent’s SureDesign tool.
- HaloPlex Custom Panel Tier 1, ION (G9902B)
- HaloPlex Target Enrichment System Kit (Agilent)
- Herculase II Fusion Enzyme with dNTPs (100 mM; 25 mM for each nucleotide) (Agilent p/n 600677)
- Agencourt AMPure XP Kit 5 mL/60 mL (Beckman Coulter Genomics p/n A63880, p/n A63881)
- Nuclease-free water (not DEPC-treated) (Ambion Cat #AM9930)
- 10 M NaOH, molecular biology grade (Sigma, p/n 72068)
- 10 mM Tris-EDTA, pH 8.0
- 2 M acetic acid (Sigma, p/n 72068)
- 100% ethanol, molecular biology grade (Sigma-Aldrich p/n E7023)
- Quant-iT dsDNA BR Assay Kit, for use with the Qubit fluorometer (100 assays, 2–1000 ng; 500 assays, 2–1000 ng) (Life technologies p/n Q32850, Life technologies p/n Q32853)
- InnuPREP Blood DNA Mini Kit (Analytik Jena AG, 845-KS-1020050)
Consumables
- 8-well PCR strip tubes with caps (Nippon Genetics p/n FG-088WF)
- Hard shell 96-well PCR plate (Eppendorf twin.tec, skirted, 0030 128.680)
- Skirted 0.5 mL tubes (STARLAB, E1405-2120)
- 2 mL tubes (STARLAB, E1420-2340)
- Tube Caps (STARLAB, 1480-0199)
- CyBio® TipBox 96/50 µL (OL 3811-25-535-N)
- CyBio® TipBox 96/250 µL (OL 3811-25-637-S)
- Qubit assay tubes (Life Technologies p/n Q32856)
Equipment
- CyBio® FeliX Head R 96/250 μL (OL3316-14-850)
- CyBio® FeliX (OL 5015-24-100) with CyBio® Composer Software
- 12-channel adapter Head R 96 (OL 3317-11-340)
- 8-channel adapter Head R 96 (OL 3317-11-330)
- Tube Rack 2.0 mL for 24 tubes (844-00136-0)
- Gripper (OL 3317-11-800)
- Magnet Adapter M96 (Biovendis, 209601)
- IsoFreeze PCR Rack SBS (LTF0045)
- BioShake 3000-T elm with PCR plate adapter (Q Instruments, 1808-0517)
- Mounting Kit BioShake 3000 Series (OL 3317-23-690)
- Reagent tube Support (OL 3317-11-240) with Reagent tube 30 mL (2320230-00)
- Waste box (844-00430-0)
- Thermal cycler 2200 TapeStation (Agilent p/n G2964AA or G2965AA)
- High Sensitivity D1000 Reagents (Agilent p/n 5067-5585)
- High Sensitivity D1000 ScreenTape (Agilent p/n 5067-5584)
- Ion PGM sequencer (Life Technologies)
- Qubit 2.0 Fluorometer (Life Technologies p/n Q32866)
- Magnet Adapter M96 (Biovendis, 209601)
- BioShake 3000-T elm with PCR plate adapter (QInstruments, 1808-0517)
- Mounting Kit BioShake 3000 Series (OL 3317-23-690)
- Waste box (844-00430-0)
- Reagent tube Support (OL 3317-11-240) with Reagent tube 30 ml (2320230-00)
- Thermal cycler 2200 TapeStation (Agilent p/n G2964AA or G2965AA)
- High Sensitivity D1000 ScreenTape (Agilent p/n 5067-5584)
- High Sensitivity D1000 Reagents (Agilent p/n 5067-5585)
- Qubit 2.0 Fluorometer (Life Technologies p/n Q32866)
- Ion PGM sequencer (Life Technologies)

Figure 1. CyBio® FeliX Liquid Handling Platform. The two-level deck design of the CyBio® FeliX possesses a small footprint while providing sufficient deck positions for reagents, accessories, and consumables
Experimental Design and Analysis
Automated library preparation was carried out on the CyBio® FeliX platform. The automation of the process adheres to the HaloPlex Target Enrichment System Kit protocol (Agilent) with a few alterations. In the course of the process, the PCR plate and tubes are not centrifuged and not cooled, apart from the Enzyme Reaction Strips on position 8 of the deck layout day 1, which was kept on a precooled PCR rack. Short incubations were carried out on deck and the PCR plate was not sealed during short incubation. Figure 2 sketches out the automation workflow. The process includes some stop points recommended in the protocol for manual steps.
Table 1. Workflow of the Agilent HaloPlex Target Enrichment System on CyBio® FeliX. Gray steps can be performed on CyBio® FeliX with minimal user intervention (add on magnetic particles). White steps are performed off-deck using additional equipment. *Automation possible; **Automation possible, but for optimal results, it is recommended to add beads manually under optical control.
| . |
| Day 1 |
Step 1 |
Digest genomic DNA with restriction enzymes |
→ |
Manually: sample preparation*
Optional: Validation of digestion |
| Step 2 |
Hybridize digested DNA to HaloPlex probe for target enrichment & sample barcoding |
→ |
Extern: PCR cycler* |
| Day 2 |
Step 3 |
Capture to target DNA |
→ |
Manually: Magnetic beads preparation** |
| Step 4 |
Ligate the captured, circulated fragments |
|
|
| Step 5 |
Prepare the PCR Mastermix |
|
|
| Step 6 |
Elute the captured DNA with NaOH |
|
|
| Step 7 |
PCR amplify the captured target libraries |
→ |
Extern: PCR cycler* |
| Step 8 |
Purify the amplified target libraries |
→ |
Manually: Magnetic beads preparation** |
| Step 9 |
Validate enrichment & qualify enriched target DNA |
→ |
Extern: 2200 TapeStation (Agilent) |
| Step 10 |
Pool samples with differnet barcodes for multiplexed sequencing |
→ |
Manually |
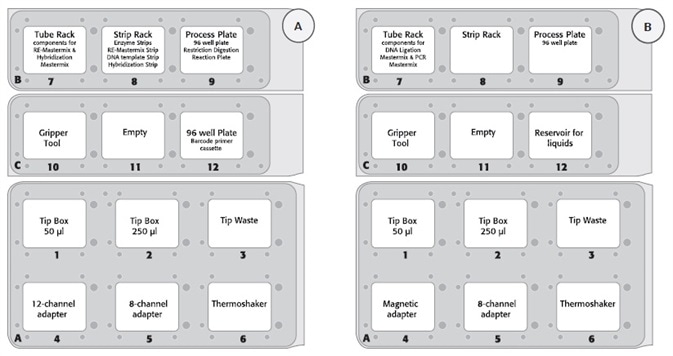
Figure 2. (A) Deck layout day 1 (protocol step 1 – 2) and (B) deck layout day 2 protocol step 3 – 8) of the CyBio® FeliX platform using Agilent HaloPlex Target Enrichment System Kit.
- Figure 2 displays the deck layout for the 2-day protocol
- The deck layout was modified due to row-wise and column-wise distribution for digestion and hybridization on day 1 and magnetic bead-based capture and purification on day 2
- To assess the automated procedure, eight patient DNA sample libraries without an internal control were prepared using the CyBio® FeliX workstation. During the automated library preparation protocol, the bead solutions for capture target DNA (HaloPlex Magnetic Beads) and purification amplified target libraries (Agencourt AMPure XP beads) were prepared and added manually. Manual bead preparation is necessary to avoid bead clumping, which results in a better homogeneous suspension and a uniform distribution of the beads on the PCR plate.
- The PCR library amplification was carried out off-deck. Amplified (sequencing ready) libraries were analyzed and measured using a High Sensitivity D1000 ScreenTape with D1000 Reagents (both Agilent) for the Agilent 2200 TapeStation.
Strip Rack, Position 8
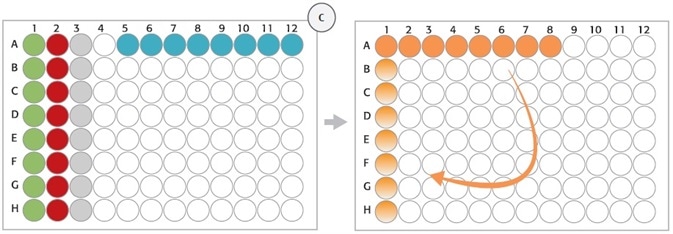
Figure 3. (C) Strip rack layout (position 8, deck layout day 1): After digestion of gDNA (step 1), Enzyme Strips (red and green), RE-Mastermix Strip (grey), and DNA template Strip (blue) are discarded and replaced by Hybridization Strip (orange). For hybridization, DNA samples were pooled in Hybridization Strip (orange). The Strip was rotated manually (light orange) before adding the HaloPlex ION Barcode Primer Cassette.
Detailed Protocol — Library Preparation
Elaborate guidelines for generating libraries with the HaloPlex Target Enrichment System Kit for Ion Torrent™ Sequencing on the CyBio® FeliX are given below.
The protocol given below contains volumes suitable for eight samples, two reaction excess. The protocol workflow with two different deck layouts is spread over two days due to a 16-hour Hybridization step.
Step 1. Digest Genomic DNA with Restriction Enzymes
Normalized genomic DNA (gDNA) samples were prepared and diluted manually and taken in an 8-well strip (Figure 2C, blue tube strip). The Restriction Enzyme Master Mix was prepared and the digestion process was carried out on the thermal shaker (BioShake 3000 T elm) on floor. The Agilent 2200 TapeStation was employed to validate the restriction digestion reaction.
Step 2. Hybridize Digested DNA to HaloPlex Probe for Target Enrichment and Sample Barcoding
HaloPlex probe was developed to hybridize selectively to target regions of the genome. 8-well strips from Step 1 (Enzyme Strips, RE-Mastermix Strip) on position 8 (columns 1–3) were manually removed and a new 8-well strip was used instead for the Hybridization Master Mix (Figure 2C, orange tube strip). The digested DNA samples from the 96-well Restriction Digest Reaction Plate were combined with the 12-channel adapter in the hybridization reaction strip. Once the DNA pooling is completed, the hybridization reaction strip had to be manually rotated prior to adding the HaloPlex ION Barcode Primer Cassette to each tube with the help of 8-channel liquid handling adapter (Figure 2C). The hybridization step was carried out overnight external in a thermal cycler using the thermal cycler program from Table 1.
Step 3. Capture the Target DNA
Manually resuspended and prepared Streptavidin-coated magnetic beads were supplied and shifted manually. For optimal results, it is recommended to add beads under optical control to guarantee that a homogenous bead suspension is distributed to all samples. The subsequent addition and cleansing steps were carried out automatically.
Step 4. Ligate the Captured, Circularized Fragments
Ligation Master Mix was prepared and mixed with the DNA captured reactions, and subsequently, incubation is performed in the on-deck thermal shaker.
Step 5. Prepare the PCR Master Mix
A PCR Master Mix was prepared for the acquired target DNA amplification step, distributed to 0.2 mL 8-well strip.
Step 6. Elute Captured DNA with NaOH
Subsequent to the ligation reaction, the acquired DNA libraries were washed with freshly prepared 50 mM NaOH.
Step 7. PCR Amplify the Captured Target Libraries
Libraries were amplified inside an external thermal cycler using the PCR Master Mix prepared in step 5. Cycling was carried out externally in a thermal cycler with parameters provided in Table 2.
Step 8. Purify the Amplified Target Libraries
AMPure XP beads were used to purify amplified DNA. The beads were prepared manually according to the recommendations in the protocol and added manually to the samples as mentioned in step 3. Amplified libraries are eluted in 10 mM Tris-EDTA and shifted into a new column of the process plate on position 9.
Step 9. Validate Enrichment and Quantify Enriched Target DNA
A 2200 TapeStation (Agilent) was used to verify size distribution of the final libraries externally. Size distribution of libraries must be between 150 and 550 bp.
Step 10. Pool Samples with Different Barcodes for Multiplexed Sequencing
This step was performed manually as recommended in the Agilent protocol.
Table 2. Probe hybridization PCR program for HaloPlex Target Enrichment for Ion Torrent™ Sequencing.
| Step |
Temperature |
Holding time |
| 1 |
95 °C |
10 min |
| 2 |
54 °C |
16 hours/overnight |
Table 3. Post-capture DNA amplification PCR program for HaloPlex Target Enrichment for Ion Torrent™ Sequencing.
| Step |
Cycle |
Temperature |
Holding time |
| 1 |
1 |
98 °C |
2 min |
| 2 |
16 (cycle number varies for each HaloPlex Probe design) |
98 °C
60 °C
72 °C |
30 sec
30 sec
1 min |
| 3 |
1 |
72 °C |
10 min |
| 4 |
1 |
8 °C |
hold |
Results and Discussion
Sample Analysis Using the 2200 TapeSation
Automated library construction was done using eight patient samples with the help of HaloPlex Target Enrichment System Kit on the CyBio® FeliX platform. To evaluate the quality of each library, the validation of enriched target DNA was analyzed using the 2200 TapeStation (Agilent) as instructed by the manufacturer. If the library has been properly compiled with adapters and barcodes to both ends of the fragment, there should be an electropherogram that displays a peak fragment size from around 150 to 550 bp. Figure 3 illustrates the TapeStation data acquired from the analysis of 2 µL of each amplified libraries. Figure 3A presents the gel image for three chosen amplified libraries (patient sample 115–117).
All three samples prepared with the HaloPlex Target Enrichment System Kit on the CyBio® FeliX were stable and had the preferred fragment size distribution, with a peak between 200 and 550 bp (Figure 3, B–D). The TapeStation-measured concentrations of the samples were more or less equivalent, 150.6 nmol/L for patient sample 115, 128.1 nmol/L for patient sample 116, and 126.3 nmol/L for patient sample 117. The yield was excellent; only 100 pmol/L is required for the sequencing process. These data present the reliable automation of NGS library preparation with the CyBio® FeliX.
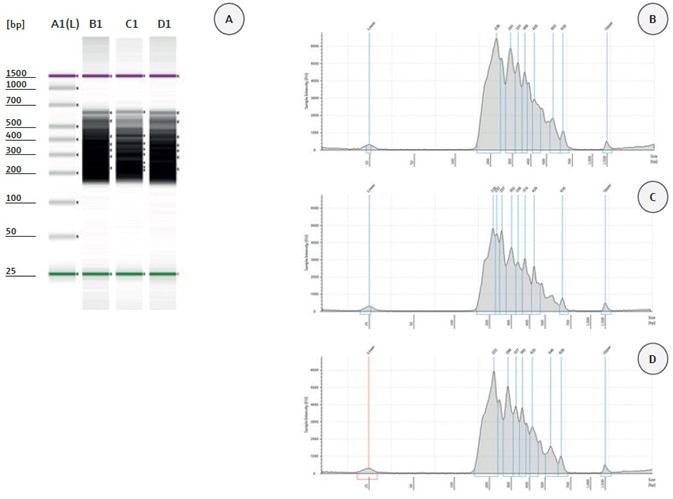
Figure 4. Validation of HaloPlex enrichment by 2200 TapeStation analysis. (A) Gel Image of 3 analyzed samples. Lane A1: High Sensitivity Ladder; Lane B1: patient sample 115; Lane C1: patient sample 116; Lane D1: patient sample117. Electropherograms (B–D): patient sample 115 (B), patient sample 116 (C), and patient sample 117 (D).
Conclusion
Automated library sample preparation for Ion Torrent™ Sequencing can be effectively carried out on the CyBio® FeliX, with the help of the HaloPlex Target Enrichment System Kit for Ion Torrent™ Sequencing. The proof of principle to produce eight high-quality libraries simultaneously was shown. Furthermore, the HaloPlex Target Enrichment System protocol from Agilent for Illumina Sequencing comprises of the same steps. The quality of the libraries prepared on the CyBio® FeliX was equivalent to that prepared manually. It is noteworthy that the application of CyBio® FeliX for automation of NGS sample preparation using the HaloPlex Target Enrichment System Kit allows easy preparation of multiple samples with less effort and greater stability.
The CyBio® FeliX is a perfect flexible liquid handling platform to enhance the reproducibility of the results and decrease the hands-on time.
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results.
This will surely ease your daily work and speed up your work processes in a certain way. All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems which dates back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness and simplicity.
Moreover, the Automation Team designs, produces and installs fully automated systems tailored to our clients' application, throughput and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems we handle your compounds, biomolecules and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.