The major histocompatibility complex (MHC), or Human Leukocyte Antigen (HLA) complex, is essential for immunorecognition, organ and stem cell transplantation, and disease association, including autoimmune disorders.1
HLA genes encode surface marker proteins for leukocyte cells. These genes are closely related and highly polymorphic, meaning they exist in various forms (alleles) in the population.
The HLA nomenclature in the IPD-IMGT/HLA database describes more than 40,000 HLA alleles. Each allele differs in nucleotide sequence, which might cause differences in the amino acid sequence of the encoded protein, affecting its function.2
HLA typing is critical for eligible transplants between recipient and donor, as HLA settings must match to reduce the likelihood of rejection from an immune reaction.
HLA typing can be performed using numerous methods, including endpoint PCR, qPCR, Sanger sequencing, and next-generation sequencing (NGS). HLA typing has grown more accessible as NGS capabilities have increased, and it now provides high-resolution analysis of target sequences.3
NGS library preparation for HLA typing uses long-range PCR amplicons that span the entire gene sequence, allowing for high-resolution typing. Long-range PCR, however, increases the probability of PCR mistakes and unspecific products due to kilobases in length.
Longer PCR products are more likely to contain GC-rich sequences, which inhibit the progression of the DNA polymerase during the PCR. The yield of long-range PCR products is typically lower than normal-length PCR, which involves several hundred base pairs.
As HLA genes are highly polymorphic, multiplex PCR is essential for typing multiple HLA loci from a single sequencing run and covering all base pair changes as precisely as possible.
Along with long-range PCR for HLA genes, allele typing using NGS is an important immunodiagnostic method.
PCR thermal cyclers are typically used for NGS sample preparation, including library preparation. The precise temperature control, efficient heating and cooling, and great temperature homogeneity across the entire block result in a consistent synthesis of amplicons and sequencing libraries free of unspecific PCR products.
Analytik Jena’s Biometra TAdvanced is a high-performance thermal cycler ideal for PCR applications such as long-range PCR and NGS library preparation.
This article outlines the HLA typing workflow with the GenDx NGSgo Library Full Kit and NGSgo-MX11-3 amplification kit. The Biometra TAdvanced 96 SG was used for all library preparation procedures, including the initial long-range PCR. Figure 1 depicts all the steps in the NGS-based HLA typing workflow.
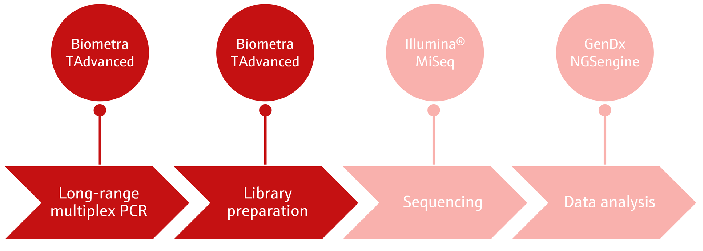
Figure 1. Overview of the NGS-based HLA typing workflow. Image Credit: Analytik Jena US
Materials and methods
Samples and reagents
- Purified DNA from blood samples or buccal swabs
- GenDx NGSgo-MX11-3, gene panel for multiplex long-range PCR
- GenDx NGSgo Library Full Kit, library preparation including random fragmentation, end-repair, adaptor binding compatible with Illumina® indexes
Instrumentation
- Biometra TAdvanced 96 SG thermal cycler, Analytik Jena
- Gel electrophoresis system, e.g. Biometra Compact Line, Analytik Jena
- Gel documentation, e.g. UVP GelStudio, Analytik Jena
- Illumina® MiSeq
Sample preparation
The concentrations of pure DNA samples ranged from 15 to 80 ng. The GenDx NGSgo-X11-3 handbook states that the PCR master mix and amplification primers for each long-range PCR mix were combined. Each mix (A, B, and C) amplifies various HLA genes (Table 1) that range in length from 2.5 to 6.7 kb.
Table 1. Amplicon length of PCR products from HLA gene loci from the GenDx NGSgo®-MX11-3 panel. Source: Analytik Jena US
| Primer panel |
Targets |
Amplicon |
Length |
| Mix A |
HLA-A |
whole gene |
3.1 kb |
| HLA-DRB1 |
whole gene* |
2.5 + 5 kb |
| HLA-DRB1 |
exon 1 |
5 kb |
| HLA-DRB3 |
whole gene* |
2.5 + 5 kb |
| HLA-DQA1 |
whole gene |
5.8 kb |
| Mix B |
HLA-B |
whole gene |
3.4 kb |
| HLA-DQB1 |
whole gene |
6.7 kb |
| HLA-DRB5 |
whole gene* |
2.6 + 4.8 kb |
| Mix C |
HLA-C |
whole gene |
3.4 kb |
| HLA-DPB1 |
exon 2-5 |
5.7 kb |
| HLA-DRB4 |
exon 2-5 |
4.3 kb |
| HLA-DPA1 |
whole gene |
5.5 kb |
* except for part of intron 1
The HLA amplicons were created using the PCR software for long-range PCR, as shown in Table 2.
Table 2. Thermal cycler program for long-range PCR of HLA loci. Source: Analytik Jena US
| Step |
Cycle |
Profile |
Temperature |
Holding time |
Ramp rates |
| 1 |
1 |
Initial denaturation |
95 °C |
3 min |
8 °C/sec |
| 2 |
30 |
Denaturation |
95 °C |
15 sec |
8 °C/sec |
| Annealing |
65 °C |
30 sec |
6 °C/sec |
| Elongation |
67 °C |
6 min |
8 °C/sec |
| 3 |
1 |
Final elongation |
67 °C |
10 min |
8 °C/sec |
| 4 |
1 |
Cooling |
15° C |
∞ |
6 °C/sec |
The PCR results from each sample mix were pooled for library construction, with a total input of 300 ng per sample. The GenDx NGSgo Library Full Kit generated the library, which included fragmentation, end-repair, adaptor ligation, and index-PCR following manufacturer specifications.
Libraries from all samples were combined and diluted to 4 nM concentration before loading onto an Illumina® flow cell. The libraries were sequenced using an Illumina® MiSeq sequencing machine with V3 chemistry.
Following sequencing, the data was processed using GenDx NGSengine software to determine the allele types of all investigated HLA genes.
Results and discussion
Long-range PCR is an important step in sample preparation for sequencing processes because the danger of unspecific amplification increases with longer PCR products.
Using the Biometra TAdvanced thermal cycler, consistent and repeatable PCR products for all HLA genes across 24 samples were produced from two independent preparation cycles (Figure 2 [a] and [b]), with no unspecific PCR products.
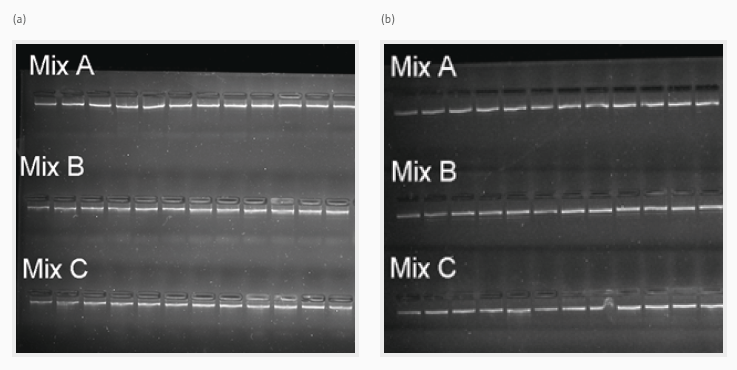
Figure 2. Gel images showing constant fragment size and stability for the long-range PCR products from all three multiplex primer combinations of two PCR runs ([a] and [b]) for following NGS library preparation and HLA gene sequencing. Image Credit: Analytik Jena US
HLA alleles were determined using the GenDx NGSengine software. Figure 3 depicts an example data overview for the two sample preparations ([a] and [b]). The sequencing reads were aligned with the reference reads.
The mappability (top panel), which indicates how many sequencing reads were allocated to the reference sequences, was high, with over 92 % across all genes.
The heterozygous locations were phased to establish the HLA type. Phasing is the process of establishing which specific alleles are paired on the same chromosome.
Figure 3 (middle panel) depicts the HLA-A gene’s depth of coverage (number of times a nucleotide was sequenced) in both samples. The average read coverage over the examined sequence was 2098 for preparation (a) and 2006 for preparation (b), indicating a high-resolution level appropriate for HLA typing.
Base variation (bottom panel) depicts the level of variability across all base positions used to identify homogeneous and heterogeneous HLA alleles. For both HLA-A genes, allele determination was best represented by a strong signal-to-noise ratio of less than 5 % base variation across all nucleotide sites.
Based on these findings, the HLA patterns of recipients and donors can be established to ensure an ideal match for transplantation with the lowest chance of rejection.
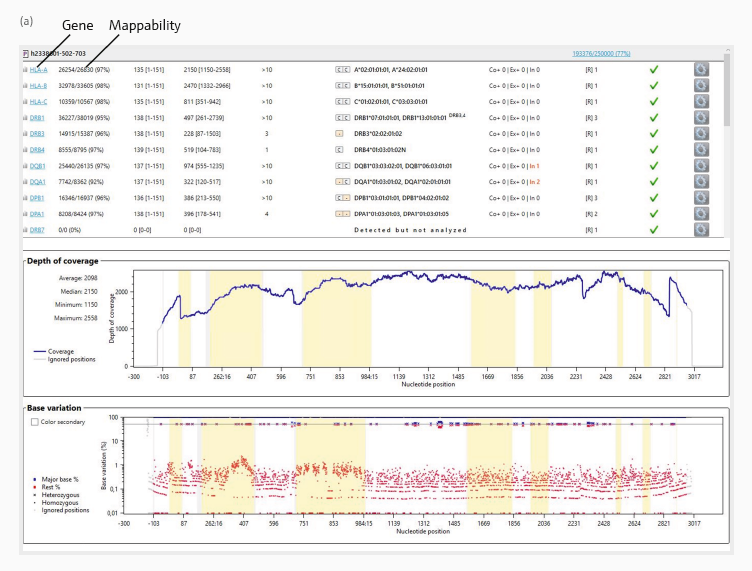
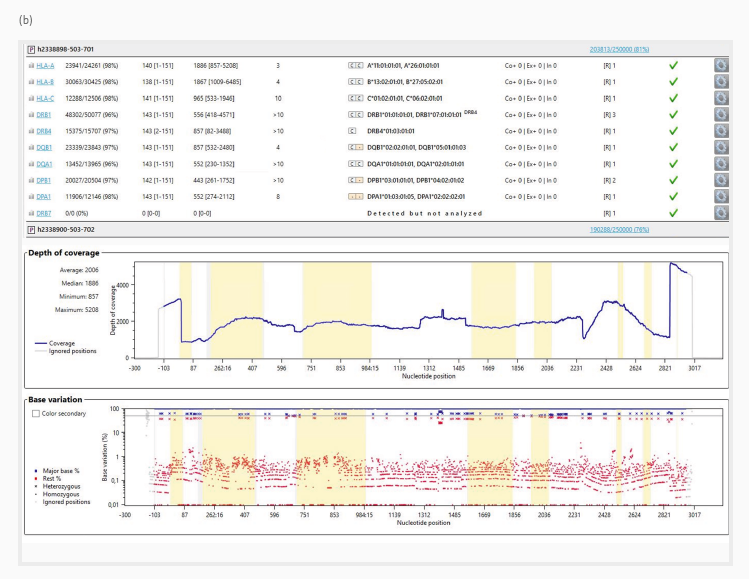
Figure 3. Two exemplary results (from preparations [a] and [b]) of HLA gene sequencing analysis using the GenDx NGSengine software. The top of both panels summarizes the analysis metrices for all sequenced genes from the two samples. The mappability for each gene to the reference sequences is displayed in the second table column.
The middle graph ‘Depth of coverage’ illustrates the number of times that a specific base was read during the sequencing process across all nucleotide positions.
The bottom graph ‘Base variation’ shows the frequency of variation in the bases at each position in the HLA gene and the determination between homozygous (dots) and heterozygous (cross) alleles. The horizontal line depicts 50 % base variation. The shaded yellow areas highlight the exon regions across the whole gene sequence.
In this example, base variation below 10 % can be classified as signal noise (red dots), while higher base variation represents heterozygous bases (red and blue crosses) or homozygous positions on the top (blue dots). Image Credit: Analytik Jena US
Summary
HLA typing is an important diagnostic method for determining the unique inventory of human cell surface antigens.
This article describes an HLA typing approach for NGS that uses Analytik Jena’s highly accurate and robust Biometra TAdvanced 96 SG thermal cycler and GenDx’s HLA qualitative in-vitro diagnostic kit.
Before library formation, samples were prepared using multiplex long-range PCR of several kilobase pairs in length for high-resolution HLA typing. Reliable and reproducible PCR products were produced without amplification bias, resulting in libraries appropriate for Illumina® sequencing.
The sequencing results demonstrated very strong mappability to reference sequences and sequence coverage for determining base variation for HLA typing, which is critical for effective organ and stem cell transplantation or immunotherapy.

Figure 4. Biometra TAdvanced 96 SG. Image Credit: Analytik Jena US
Recommended device configuration
Table 3. Thermal cycler for amplification and library preparation and electrophoresis instruments for fragment size validation. Analytik Jena US
| Article |
Article number |
Description |
| Biometra TAdvanced 96 SG |
846-x-070-241 |
End-point PCR thermal cycler |
| Biometra Compact Series |
846-025-zzz |
Horizontal gel electrophoresis family for gel
sizes from mini to maxi gels and from low to high sample throughput |
x = 2 for 230 V, x = 4 for 115 V, x = 5 for 100 V
zzz = different sizes
Acknowledgments
The data was provided in collaboration with the Laboratory of Transplant Immunology and Molecular Genetics at Jena University Hospital.
References
- Mayor, N. P. et al. HLA Typing for the Next Generation. PLOS ONE 10, e0127153 (2015).
- Buhler, S. & Sanchez-Mazas, A. HLA DNA Sequence Variation among Human Populations: Molecular Signatures of Demographic and Selective Events. PLOS ONE 6, e14643 (2011).
- Bravo-Egana, V., Sanders, H. & Chitnis, N. New challenges, new opportunities: Next generation sequencing and its place in the advancement of HLA typing. Human Immunology 82, 478–487 (2021).
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results. This will surely ease your daily work and speed up your work processes in a certain way.
All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems that date back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters, and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high-quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness, and simplicity.
Moreover, the Automation Team designs produce, and install fully automated systems tailored to our clients' application, throughput, and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems, we handle your compounds, biomolecules, and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.