Automated library preparation is critical for reducing errors, increasing repeatability, and reducing hands-on time, allowing researchers to generate sequence data from DNA more quickly.
The CyBio FeliX, a benchtop instrument for automated liquid handling tasks, was used to automate the HaloPlex Target Enrichment System Kit for NGS library preparation for Ion Torrent™ Sequencing. This resulted in reliable and highly accurate libraries for downstream sequencing.
This article demonstrates how to successfully produce high-quality DNA libraries on CyBio FeliX for personal genome analysis using the Ion Torrent™ Sequencing System.
Analytik Jena’s liquid handling portfolio includes CyBio FeliX, a compact and adaptable liquid-handling platform for a wide range of automated operations. The system offers 12 deck locations on two levels for microplates, tube racks, reservoirs, and tips with the smallest footprint possible, including attachments such as a gripper, magnet adaptor, and thermal shaker.
Library preparation is an important, hands-on, and time-consuming stage in the Next Generation Sequencing (NGS) process. Analytik Jena created a procedure for the HaloPlex Target Enrichment System Kit. It required minimal user intervention to prepare eight library samples for multiplexed sequencing on the Ion Torrent™ platform.
The Institute for Laboratory Medicine at the Donauspital in Vienna experimented with the CyBio FeliX liquid handling platform to create DNA sample libraries from patient samples.
Target enrichment of genomes improves the efficiency of NGS by allowing researchers to focus on specific regions of interest. The kit is ideal for deep sequencing of small gene panels (up to 2.5 Mb), such as those used in studies of hereditary disorders, cancer, and infectious and neurologic diseases.
Materials and methods
Consumables
- Hard-shell 96-well PCR plate (Eppendorf twin.tec, skirted, 0030 128.680)
- 8-well PCR strip tubes with caps (Nippon Genetics FG-088WF)
- 2 mL STARLAB tubes (E1420-2340)
- Skirted 0.5 mL STARLAB tubes (E1405-2120)
- Tube Caps: STARLAB (1480-0199)
- CyBio FeliX TipBox, 96/250 µL (OL 3811-25-637-S)
- CyBio FeliX TipBox, 96/50 µL (OL 3811-25-535-N)
- Qubit assay tubes (Life Technologies, p/n Q32856)tur
Kits and reagents
- To prepare the library for Ion Torrent™ sequencing, eight normalized patient DNA samples (225 ng) were extracted using the innuPREP Blood DNA Mini Kit (IST Innuscreen GmbH). The Qubit fluorometer (Life Technologies) was used to quantify the patient’s DNA. Each sample (patient DNA) to be sequenced received a unique target-enriched, barcoded library. The HaloPlex probes were created using Agilent’s SureDesign tool
- Agilent HaloPlex Target Enrichment System Kit
- HaloPlex Custom Panel Tier 1 (ION) (G9902B)
- Agencourt AMPure XP Kit (5 mL / 60 mL) (Beckman Coulter Genomics p/n A63880, p/n A63881)
- Herculase II Fusion Enzyme with dNTPs (100 mM; 25 mM for each nucleotide) (Agilent p/n 600677)
- Ambion Cat #AM9930: Nuclease-free water (not DEPC treated)
- 10 M NaOH, molecular biology grade (Sigma, p/n 72068)
- 2 M acetic acid (Sigma, p/n 72068)
- 10 mM Tris-EDTA (pH 8.0)
- Sigma-Aldrich p/n E7023: 100 % ethanol for molecular biology
- deltaPREP Blood DNA Mini Kit (IST Innuscreen GmbH, 31-DP-100050)
- Quant-iT dsDNA BR Assay Kit for use with the Qubit fluorometer (100 assays, 2‒1000 ng and 500 assays, 2‒1000 ng) (Life Technologies p/n Q32850, Q32853)
Equipment
- CyBio FeliX (OL 5015-24-100) with CyBio Composer Software
- CyBio FeliX Head R, 96/250 µL (OL3316-14-850)
- Eight-channel adapter Head R 96 (OL 3317-11-330)
- 12-channel adapter Head R 96 (OL 3317-11-340)
- Gripper (OL 3317-11-800)
- Tube Rack 2.0 mL for 24 tubes (844-00136-0)
- IsoFreeze PCR Rack SBS (LTF0045)
- Biovendis Magnet Adapter M96 (209601)
- BioShake 3000-T elm with PCR plate adapter (QInstruments, 1808-0517)
- Mounting Kit BioShake 3000 Series (OL 3317-23-690)
- Waste box (844-00430-0)
- Reagent tube support (OL 3317-11-240) and 30 mL reagent tube (2320230-00)
- Thermal cycler 2200 TapeStation (Agilent p/n G2964AA or G2965AA)
- High sensitivity D1000 ScreenTape (Agilent p/n 5067-5584)
- High sensitivity D1000 reagents (Agilent p/n 5067-5585)
- Qubit 2.0 Fluorometer (Life Technologies, model Q32866)
- Ion PGM sequencer (Life Technologies)

Figure 1. CyBio FeliX Liquid Handling Platform. The two-level deck design of the CyBio FeliX possesses a small footprint while providing sufficient deck positions for reagents, accessories and consumables. Image Credit: Analytik Jena US
Experimental design and analysis
The CyBio FeliX platform was used to complete automated library preparation. The procedure was automated using the HaloPlex Target Enrichment System Kit methodology (Agilent), with a few alterations.
Except for the Enzyme Reaction Strips on position 8 of the deck layout on day one, which were placed on a precooled PCR rack, the PCR plate and tubes were neither centrifuged nor cooled during the procedure. Short incubations were performed on deck, with the PCR plate not sealed.
The automated procedure is depicted in Figure 2. The approach includes stop points specified by the protocol for manual steps.
Table 1. Workflow of the Agilent HaloPlex Target Enrichment System on CyBio FeliX. Gray steps can be performed on CyBio FeliX with minimal user intervention (add on magnetic particles). White steps are performed off-deck using additional equipment. Source: Analytik Jena US
| |
|
|
|
|
| Day 1 |
Step 1 |
Digest genomic DNA with restriction enzymes |
→ |
Manually: sample preparation* Optional: Validation of digestion |
| Step 2 |
Hybridize digested DNA to HaloPlex probe
for target enrichment & sample barcoding |
→ |
Extern: PCR cycler* |
| Day 2 |
Step 3 |
Capture to target DNA |
→ |
Manually: Magnetic beads preparation** |
| Step 4 |
Ligate the captured, circulated fragments |
|
|
| Step 5 |
Prepare the PCR Mastermix |
|
|
| Step 6 |
Elute the captured DNA with NaOH |
|
|
| Step 7 |
PCR amplify the captured target libraries |
→ |
Extern: PCR cycler* |
| Step 8 |
Purify the amplified target libraries |
→ |
Manually: Magnetic beads preparation** |
| Step 9 |
Validate enrichment & qualify enriched target DNA |
→ |
Extern: 2200 TapeStation (Agilent) |
| Step 10 |
Pool samples with differnet barcodes
for multiplexed sequencing |
→ |
Manually |
*Automation possible; **Automation possible, but for optimal results we recommended to add beads manually under optical control.
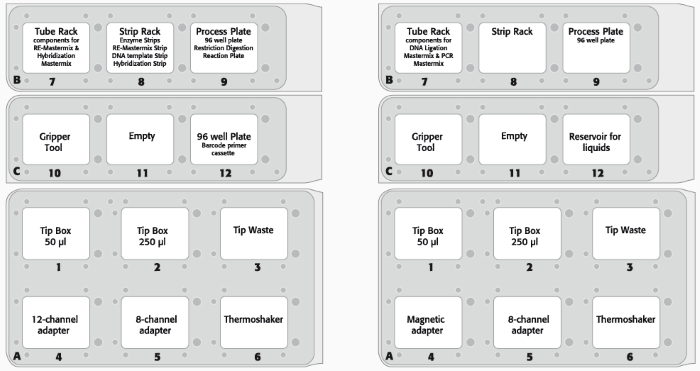
Figure 2. (A) Deck layout day 1 (protocol step 1–2) and (B) deck layout day 2 (protocol step 3–8) of the CyBio FeliX platform using Agilent HaloPlex Target Enrichment System Kit. Image Credit: Analytik Jena US
- Figure 2 shows the deck configuration for the two-day technique.
- The deck arrangement was modified to account for the distribution of digestion and hybridization on day one and magnetic bead-based capture and purification on day two.
- Using the CyBio FeliX workstation, eight patient DNA sample libraries were created without internal control to evaluate the automated technique. Throughout the automated library preparation technique, bead solutions for capturing target DNA (HaloPlex Magnetic Beads) and purifying amplified target libraries (Agencourt AMPure XP beads) were organized and added by hand. Manual bead preparation prevents clumping, resulting in a more uniform suspension and dispersion of beads on the PCR plate.
- The PCR library amplification was completed off-deck. Amplified (sequencing ready) libraries were analyzed and quantified using an Agilent High Sensitivity D1000 ScreenTape and D1000 Reagents for the Agilent 2200 TapeStation.
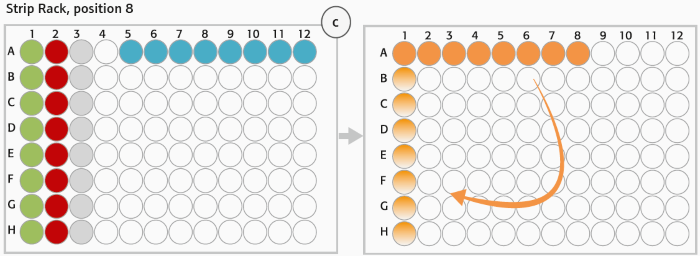
Figure 3. (C) Strip rack layout (position 8, deck layout day 1): After digestion of gDNA (step 1) Enzyme Strips (red and green), RE-Mastermix Strip (grey) and DNA template Strip (blue) are discarded and replaced by Hybridization Strip (orange). For hybridization DNA samples were pooled in Hybridization Strip (orange). The Strip was rotated manually (light orange) before adding the HaloPlex ION Barcode Primer Cassette. Image Credit: Analytik Jena US
Detailed protocol ‒ Library preparation
The following instructions detail the creation of libraries using the HaloPlex Target Enrichment System Kit for Ion Torrent™ sequencing on the CyBio FeliX. Please refer to the protocol for any instructions not contained in this article.
The protocol below includes volumes for eight samples and two reaction excess. The protocol process with two different deck configurations is extended over two days due to a 16-hour hybridization stage.
Step 1: Digest genomic DNA with restriction enzymes.
Normalized genomic DNA samples were manually produced, diluted, and placed in an 8-well strip (Figure 2C, blue tube strip).
The Restriction Enzyme Master Mix was manufactured, and the digestion process was carried out on the thermal shaker (BioShake 3000 T elm) on deck. Agilent 2200 TapeStation was used to validate the restriction digestion process.
Step 2: Hybridize digested DNA to HaloPlex probe for target enrichment and sample barcoding.
The HaloPlex probe was developed to hybridize specific areas of the genome. The 8-well strips from Step 1 (Enzyme Strips, RE-Mastermix Strip) at position 8 (columns 1–3) were manually discarded and replaced with a new 8-well strip for the Hybridization Master Mix (Figure 2C, orange tube strip).
The digested DNA samples from the 96-well Restriction Digest Reaction Plate were combined with the 12-channel adaptor in the hybridization reaction strip.
After DNA pooling, the hybridization reaction strip has to be manually rotated before introducing the HaloPlex ION Barcode Primer Cassette to each tube via the eight-channel liquid handling adapter (Figure 2C). The hybridization stage was carried out overnight in an external heat cycler with the thermal cycler protocol from Table 1.
Step 3: Capture the target DNA.
Manually resuspended and prepared Streptavidin-coated magnetic beads were provided and transferred manually. For the best results, adding beads under optical control is recommended to ensure a uniform bead suspension is disseminated over all samples. The following mixing and washing stages were carried out automatically.
Step 4: Ligate the captured, circularized fragments.
The Ligation Master Mix was made and applied to the DNA capture reactions, which were then incubated in the on-deck thermal shaker.
Step 5: Prepare the PCR Master Mix.
A PCR Master Mix was made for the captured target DNA amplification phase and dispersed to a 0.2 mL 8-well strip.
Step 6. Elute captured DNA with NaOH.
After the ligation procedure, the collected DNA libraries were eluted using freshly produced 50 mM NaOH.
Step 7: PCR amplify the captured target libraries.
Libraries were amplified in an external thermal cycler using the PCR Master Mix created in Step 5. Cycling was done externally in a thermal cycler under the conditions in Table 2.
Step 8: Purify the amplified target libraries.
Amplified DNA was purified with AMPure XP beads. The beads were produced as directed by the technique and manually added to the samples as described in Step 3. Amplified libraries are eluted in 10 mM Tris-EDTA and transferred to a fresh column of the process plate at position 9.
Step 9: Validate enrichment and quantify enriched target DNA.
The size distribution of the resulting libraries was checked externally with an Agilent 2200 TapeStation. The size distribution of libraries should be between 150 and 550 bp.
Step 10: Pool samples with different barcodes for multiplexed sequencing.
This step was completed manually following the Agilent protocol.
Table 2. Probe hybridization PCR program for HaloPlex Target Enrichment for Ion Torrent™ Sequencing. Source: Analytik Jena US
| Step |
Temperature |
Holding time |
| 1 |
95° C |
10 min |
| 2 |
54° C |
16 hours/overnight |
Table 3. Post-capture DNA amplification PCR program for HaloPlex Target Enrichment for Ion Torrent™ Sequencing. Source: Analytik Jena US
| Step |
Cycle |
Temperature |
Holding time |
| 1 |
1 |
98 °C |
2 min |
| 2 |
16 (cycle number
varies for each
HaloPlex Probe
design) |
98 °C |
30 sec |
| 60 °C |
30 sec |
| 72 °C |
1 min |
| 3 |
1 |
72 °C |
10 min |
| 4 |
1 |
8 °C |
hold |
Results and discussion
Sample analysis using the 2200 TapeSation
The 2200 TapeSation was used to analyze eight patient samples and create an automated library utilizing the CyBio FeliX platform’s HaloPlex Target Enrichment System Kit.
To evaluate the quality of each library, the validation of enriched target DNA was performed using the Agilent 2200 TapeStation following manufacturer instructions.
If the library was appropriately constructed, with adapters and barcodes at both ends of the fragment, the electropherogram should indicate a peak fragment size of 150 to 550 bp. Figure 3 displays TapeStation data received from analyzing 2 µL of each amplified library.
Figure 3A shows the gel picture for three selected amplified libraries (patient samples 115–117). All three samples generated with the CyBio FeliX using the HaloPlex Target Enrichment System Kit were consistent and exhibited the expected fragment size distribution, with a peak between 200–550 bp (Figure 3, B–D).
The TapeStation-measured concentrations of the samples were nearly identical: 150.6 nmol/L for patient sample 115, 128.1 nmol/L for patient sample 116, and 126.3 nmol/L for patient sample 117.
The yield was excellent, with only 100 pmol/L required for the sequencing process. These results show that the CyBio FeliX can reliably automate NGS library preparation.
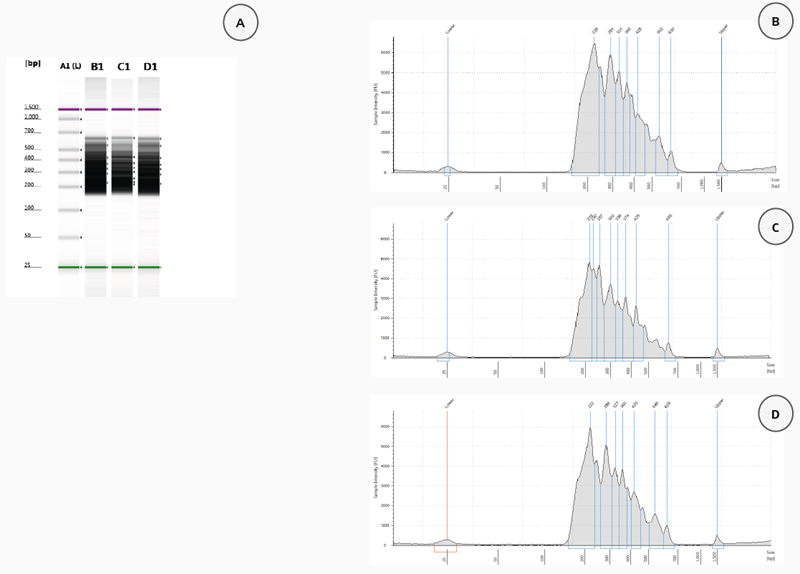
Figure 4. Validation of HaloPlex enrichment by 2200 TapeStation analysis. (A) Gel Image of 3 analysed samples. Lane A1: High Sensitivity Ladder, Lane B1: patient sample 115, Lane C1: patient sample 116, Lane D1: patient sample 117. Electropherograms (B-D): patient sample 115 (B), patient sample 116 (C), patient sample 117 (D). Image Credit: Analytik Jena US
Summary
The CyBio FeliX successfully prepares library samples for Ion Torrent™ sequencing using the HaloPlex Target Enrichment System Kit. A proof of concept demonstrated the ability to build eight high-quality libraries in parallel.
Furthermore, the HaloPlex Target Enrichment System methodology from Agilent for Illumina Sequencing follows the same stages. The quality of libraries prepared with the CyBio FeliX was comparable to those prepared manually.
It should be noted that using CyBio FeliX to automate NGS sample preparation with the HaloPlex Target Enrichment System Kit allows for the easy preparation of numerous samples with less work and more uniformity.
The CyBio FeliX is a perfect flexible liquid handling device for increasing reproducibility and reducing hands-on time.
Acknowledgments
Dr. A. Robubi of the Donauspital Institute for Laboratory Medicine, Langobardenstr. 122, A-1220 Vienna, Austria, created and kindly provided the data described in this article.
References
- Reference: AN_0439_0024_en_1605.docx
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results. This will surely ease your daily work and speed up your work processes in a certain way.
All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems that date back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters, and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high-quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness, and simplicity.
Moreover, the Automation Team designs produce, and install fully automated systems tailored to our clients' application, throughput, and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems, we handle your compounds, biomolecules, and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.