Human leukocyte antigen (HLA) genotyping is critical when preparing for stem cell transplants. The procedure is crucial to determine the genetic compatibility of the HLAs between the donor and recipient.1 A high level of compatibility considerably minimizes the risk of rejection and improves transplant success.
Next-generation sequencing (NGS) provides high-resolution, exact identification of HLA alleles, making it a valuable tool in stem cell transplantation.2 Accurate HLA genotyping is vital for effective results in this process.
Automating sample preparation for HLA genotyping has various benefits, including enhanced efficiency, lower error rates, and reduced manual handling time. The cleanup phase is critical during the NGS library preparation process.
This step entails refilling the buffer containing DNA fragments and eliminating residual enzymes and components from earlier preparation phases, such as primers and adapters. Several methods have been developed for DNA fragment cleanup, with magnetic beads being the most simple.
Magnetic beads allow DNA molecules to bind with their unique coating and optimal buffer conditions. The size of the bead-bound DNA fragments is largely determined by the concentration of the buffer components in which the beads are kept.
The bead-to-sample ratios can be adjusted to preferentially extract DNA fragments within a user-defined size range. In this case, the liquid handling robot CyBio FeliX (Figure 1) was used to remove fragments ranging in size from 500 to 800 bp for sequencing.

Figure 1. Analytik Jena CyBio FeliX hardware configuration. Decks equipped for doublesided size selection of NGS libraries. Image Credit: Analytik Jena US
Materials and methods
Reagents and consumables
- NGSgo® Library Full Kit (GenDx)
- NGSGo® Ampx v2 HLA-A, B, C, DRB1, DQB1, DPB1 (GenDx)
- AMPure XP Reagent (A63881, Beckman Coulter)
- Freshly prepared 80 % Ethanol (9065.3, Carl Roth GmbH &CO.KG)
- Freshly prepared 0.1x Tris-EDTA buffer (93283-100ML, Sigma Aldrich)
- 96-deep well plate as a waste reservoir, 12-column reservoir for beads storage, 2x single-well reservoir
- The 96-well PCR plate containing DNA fragments
- 2x 96 well PCR plate
- Optional for non-skirted and half-skirted 96-well PCR plates: 2x Adapter 96, passive cooling function (844-00135-0, Analytik Jena)
- CyBio RoboTipTray 96/250 µL; DW PCR-certified, pre-sterilized, filter (OL3810-25-669, Analytik Jena)
- Optional: Protective Plate (OL3317-25-125, Analytik Jena)
- dsDNA BR Assay (Q32850, Thermo Fisher Scientific)
- QInstruments BioShake 3000-T elm (QINSTRUMENTS-2016-0517, Analytik Jena)
- QINSTRUMENTS Adapter for 96/200 µL Bio-Rad PCR plate (848-2016-1042, Analytik Jena)
- Mounting Kit; BioShake 3000 Series (OL3317-23-692, Analytik Jena)
- ALPAQUA® MAGNUM FLX™; Universal Magnet Adapter (OL3317-11-285, Analytik Jena)
- Gripper (OL3317-14-800, Analytik Jena)
- Support; 70 mm height (OL3317-11-110, Analytik Jena)
- Endpoint thermal cycler, e.g. Biometra T Advanced 96 S (846-x-070-251, x = 2 for 230 V, 4 for 115 V, 5 for 100 V, Analytik Jena)
- InvitrogenTM QubitTM3 (Thermo Fisher Scientific)
- NovaSeq (Illumina®)
Software
- CyBio Composer Software (Analytik Jena)
- NGSengine software (GenDx)
Methods on CyBio FeliX
The approach proposed here automates the size selection of DNA libraries for NGS with magnetic beads utilizing the CyBio FeliX liquid handling platform (Figure 1).
The automated stages were integrated into the library preparation process of the NGSgo® Library Full Kit (GenDx) for NGS HLA typing. Before library preparation, the NGSGo® Ampx v2 kit (GenDx) was used to perform singleplex PCR on six distinct HLA loci.
Following PCR amplification, DNA quantities were determined using a Qubit assay, and all PCR products were pooled for each sample. The library preparation was continued following the manufacturer’s instructions.
After adapter ligation, an automated library cleanup was completed. The size selection was performed following the indexing PCR, utilizing the deck arrangement shown in Figure 2.
Figure 3 illustrates the entire procedure, including each phase of fragment size selection and library purification. Steps 1-14 are necessary only when performing double-sided size selection. For amplified library cleanup, which focuses on removing small sample fragments, steps 4-8 are omitted.
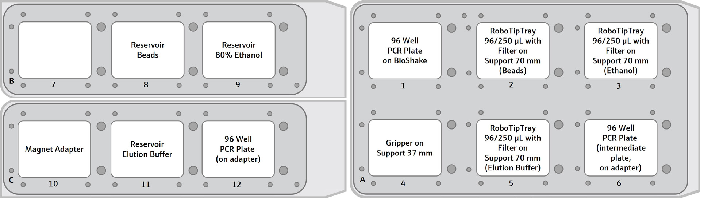
Figure 2. Graphical deck layout for Protocol – Double-sided Size Selection. The CyBio FeliX provides 12 deck positions on 3 movable decks. The upper decks B and C equipped with the required accessories are shown on the left, the lower deck A is shown on the right. The 12-column reservoir on position 8 is equipped with 2 mL AMPure XP beads per column. Image Credit: Analytik Jena US
The CyBio FeliX carried out the size selection stages outlined in the instructions below:
Step 1: Setup of large fragment removal
Each well of the PCR plate on the BioShake receives 10.4 µL of AMPure XP beads directly from the reservoir. The plate already has 26 µL of DNA fragments per well combined with beads by pipetting up and down five times. The remaining beads in the tips are dumped into a 96-deep trash plate.
Step 2: Incubation DNA collection
Incubate the beads and DNA fragments for five minutes at room temperature and combine at 1600 rpm. Note: After incubation, the beads should be uniformly distributed throughout the solution.
Step 3: Bead separation
Using the gripper, place the PCR plate containing DNA fragments and beads onto the magnet adapter. Incubate at room temperature for five minutes, ensuring that no beads remain suspended. If beads are still suspended, extend the incubation period as needed.
Step 4: Supernatant transfer
In a single pipetting step, transfer the supernatant containing unbound DNA fragments into a fresh 96-well PCR plate using a RoboTipTray 96/250 µL. Place the plate on the BioShake using the gripper.
Step 5: Tip exchange
Manually replace the tips used for bead dispersion and supernatant transfer.
Step 6: Setup for bead cleanup
From the beads reservoir, 3.9 µL are transferred into each well of the 96 well PCR plate containing the supernatant. The beads are mixed with the DNA containing solution by pipetting five times.
Step 7: Second incubation
Incubate and mix the DNA collection beads and fragments for five minutes at room temperature and 1600 rpm. Note: After incubation, the beads should be evenly dispersed.
Step 8: Second bead separation
Using the gripper, place the PCR plate containing DNA fragments attached to beads onto the magnet adapter and incubate at room temperature for five minutes. After incubation, no beads should remain in the solution.
Step 9: Discard supernatant
Using a RoboTipTray 96/250 µL, pipette the supernatant into the waste plate in a single step.
Step 10: Ethanol wash
Add 100 µL 80 % ethanol to the beads using a new RoboTipTray 96/250 µL. The beads are incubated with ethanol for 30 seconds while remaining on the well’s wall.
After incubation, the supernatant is removed using the tips previously used for bead transfer. The supernatant is then discarded in the waste plate. This ethanol wash is repeated another two times.
Step 11: Bead drying
Incubate the beads for three minutes at room temperature to allow any residual ethanol to evaporate.
Step 12: DNA fragment elution
Place the PCR plate containing dried beads onto the BioShake using the gripper. With new tips, add 25 µL of elution buffer (0.1x Tris-EDTA) from the reservoir to the dried beads. To ensure proper resuspension, pipette the DNA fragments five times, then shake the plate for five minutes at 1600 rpm.
Step 13: Final bead separation
Place a PCR plate containing resolved beads on the magnet adapter using the gripper and incubate at room temperature for five minutes. Beads should not remain in the solution after incubation.
Step 14: Elution buffer transfer
Transfer 20 µL of the supernatant containing the target-size DNA fragments into a fresh PCR plate.
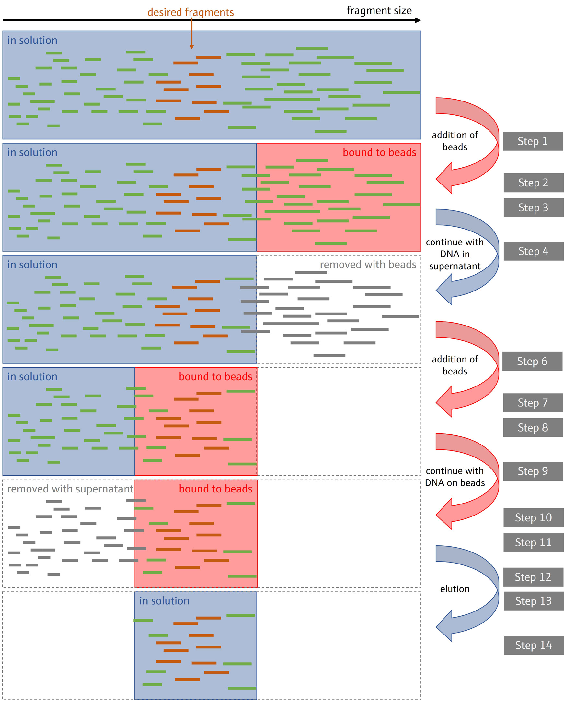
Figure 3. Illustration of the size selection process. Double-sided size selection in NGS library preparation using magnetic beads to selectively bind and isolate DNA fragments within a desired size range.
By performing two rounds of bead cleanup with different bead-to-sample ratios, fragments outside the target size range are effectively removed, ensuring a library of appropriately sized DNA fragments for sequencing. Image Credit: Analytik Jena US
Results and discussion
During library construction, DNA concentration measurements were taken before and after size selection and fragment size evaluations after sequencing using the NGSengine software (GenDx) to analyze the effectiveness of the size selection processes.
The fall in DNA concentration following the 0.7x bead clean-up and size selection procedures demonstrates that undesired DNA fragments and impurities were successfully removed.
The 0.7x bead cleanup resulted in significant decreases in average DNA concentration (t-test, p > 0.05), ranging from -4.4 to -34.6 ng/µL across all samples, indicating efficient purification via the automated procedure (Figure 4).
Similarly, size selection resulted in a constant drop in DNA concentration, ranging from -2.4 to -15.4 ng/µL, targeting the isolation of DNA fragments within the targeted size range (600–800 base pairs).
These findings demonstrate that the cleansing and size selection operations effectively enrich the samples for high-quality, properly sized DNA fragments required for good sequencing performance.
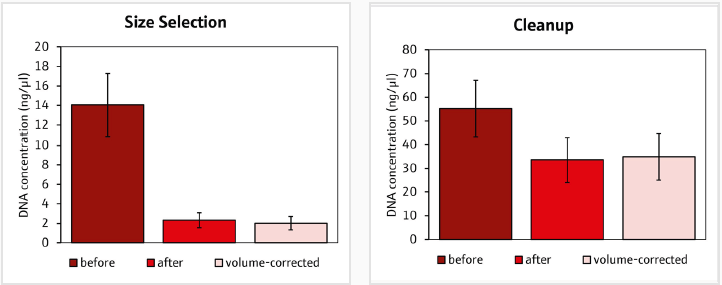
Figure 4. DNA concentrations of sequencing libraries before and after the A) size selection and B) clean up. Volume corrected concentrations were based on different elution volumes after the size selection and clean up. Image Credit: Analytik Jena US
The automated procedure resulted in considerably larger fragment sizes (t-test, p < 0.05), ranging from 250 bp to over 350 bp, compared to the manual process, which ranged from 210 bp to 320 bp. This aligns with the purpose of size selection (Figure 5).
As a result, automating processes ensures the reliable and efficient selection of optimally sized fragments for subsequent sequencing procedures. The advantage of the automated method over the manual process on CyBio FeliX is that all samples can be processed concurrently without delay.
This prevents tiny particles from attaching too tightly to the beads. Accurate adherence to incubation durations guarantees that the fragments are correctly sized.
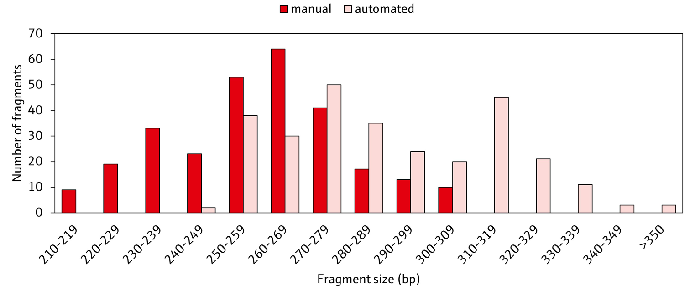
Figure 5. Comparison of the average sequence fragment sizes after manual and automatic size selection with CyBio FeliX. The average fragment sizes of six HLA loci from 48 samples are displayed after trimming of sequencing adapters and barcodes using the NGSengine software (GenDx).
Fragment sizes in the automated workflow (light red) were generally larger compared to the manual process (dark red), which grants higher coverage to reference sequences and higher resolution for HLA typing. Image Credit: Analytik Jena US
Optimal sequencing conditions are achieved through accurate size selection, ensuring ideal sequencing results. The automated system is more efficient, performing multiple cleanups sequentially, which accelerates the process compared to manual methods.
Another advantage is the increased precision, as manual procedures are more prone to errors, while automation enhances reproducibility. Additionally, the ergonomics of automation reduce the physical strain associated with manual handling, making the process more user-friendly.
The automated method, which consistently produces bigger DNA fragments and effectively handles multiple samples, not only meets sequencing requirements but also provides considerable benefits from throughput, reproducibility, and error reduction.
This ensures the best conditions for sequencing and improves overall process efficiency.
Summary
The CyBio FeliX allows for the simultaneous processing of 96 samples in approximately 40 minutes, with the flexibility to perform multiple runs throughout the day. Its compact design reduces the bench space needed for automated library preparation, optimizing sequencing on an Illumina® NovaSeq.
Using the CyBio FeliX accelerates these processes by a factor of four compared to manual methods and significantly increases walk-away time for automation.

Figure 6. CyBio FeliX. Image Credit: Analytik Jena US
Recommended device configuration
Table 1. Overview of devices, accessories, and consumables. Source: Analytik Jena US
| CyBio FeliX Accessoires |
Order number |
| CyBio FeliX Basic Unit with Enclosure |
OL5015-24-100 |
| CyBio FeliX Head R 96/250 μL |
OL3316-14-850 |
| QInstruments BioShake 3000-T elm |
QINSTRUMENTS-2016-0517 |
| QINSTRUMENTS Adapter for 96/200 μL Bio-Rad PCR plate |
848-2016-1042 |
| Mounting Kit; BioShake 3000 Series |
OL3317-23-692 |
| ALPAQUA® MAGNUM FLX™, Universal Magnet Adapter |
OL3317-11-285 |
| Gripper |
OL3317-14-800 |
| Support, 70 mm, 3x |
OL3317-11-110 |
| CyBio RoboTipTray 96/250 μL DW; PCR-certified, pre-sterilized, filter |
OL3810-25-669 |
| Optional: |
|
| Adapter 96, passive cooling function, 2x |
844-00135-0 |
| Protective Plates |
OL3317-25-125 |
Acknowledgments
The authors thank Eugen Bauer, M.Sc. (Transfusion Medicine, University of Cologne), for his outstanding contributions.
References
- Mayor, N. P. et al. HLA Typing for the Next Generation. PLOS ONE 10, e0127153 (2015)
- Bravo-Egana, V., Sanders, H. & Chitnis, N. New challenges, new opportunities: Next generation sequencing and its place in the advancement of HLA typing. Human Immunology 82, 478–487 (2021)
About Analytik Jena US
Analytik Jena is a provider of instruments and products in the areas of analytical measuring technology and life science. Its portfolio includes the most modern analytical technology and complete systems for bioanalytical applications in the life science area.
Comprehensive laboratory software management and information systems (LIMS), service offerings, as well as device-specific consumables and disposables, such as reagents or plastic articles, complete the Group’s extensive range of products.
About Life Science
The Life Science product area demonstrates the biotechnological competence of Analytik Jena AG. We provide a wide product spectrum for automated total, as well as individual solutions for molecular diagnostics. Our products are focused to offer you a quality and the reproducibility of your laboratory results. This will surely ease your daily work and speed up your work processes in a certain way.
All together we support you through the complete process of the lab work. Besides we offer customized solutions and are able to adapt our products to your needs. Automated high-throughput screening systems for the pharmaceutical sector are also part of this segment’s extensive portfolio.
About Analytical Instrumentation
Analytik Jena has a long tradition in developing high-performance precision analytical systems that date back to the inventions made by Ernst Abbe and Carl Zeiss. We have grown to become one of the most innovative manufacturers of analytical measuring technology worldwide.
Our business unit Analytical Instrumentation offers excellent competencies in the fields of optical spectroscopy, sum parameters, and elemental analysis. Being proud of our core competency we grant all our customers a long-term warranty of 10 years for our high-performance optics.
About Lab Automation
With more than 25 years of market experience, Analytik Jena with its CyBio® Product Line is a leading provider for high-quality liquid handling and automation technologies. In the pharmaceutical and life science industries, our products enjoy the highest reputation for precision, reliability, robustness, and simplicity.
Moreover, the Automation Team designs produce, and install fully automated systems tailored to our clients' application, throughput, and capacity requirements. From stand-alone CyBio® Well up to fully customized robotic systems, we handle your compounds, biomolecules, and cells with great care.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.