Introduction
Induced pluripotent stem cells (iPSCs) are characterized by the ability to self-renew and retain their pluripotency just like embryonic stem cells. As a result, they have the ability to be distinguished into any cell type of the body, making them ideal candidates for a variety of applications, including regenerative medicine, drug screening, and toxicology.
Since the publication of breakthrough results from Dr. Yamanaka’s laboratories on the creation of mouse [1] and human [2] induced pluripotent stem cells from completely differentiated cells, research labs have been focusing on the development of an efficient approach of deriving these cells, without leaving behind a genetic footprint as a byproduct from the technique used to produce them.
Existing protocols include lentivirus and adenovirus transduction as techniques to introduce various transcription factors into terminally differentiated cells so that they can be dedifferentiated back to the pluripotent state of the iPSC [3-5].
The most commonly used combination of transcription factors, typically called the Yamanaka Factors, include a mixture of c-MYC, KLF4, SOX2 and OCT4 [1,2]. However, several weeks are required to complete these protocols, and only a small number of pluripotent clones are produced.
These drawbacks make successful reprogramming a rare event. More recently, induced pluripotent stem cells have been generated using a RNA virus called Sendai [6].
This virus reproduces in the host cell’s cytoplasm and does not attach to the host genome. It infects various types of cells, enabling the reprogramming of cells of several different linages.
Also, the Sendai system has been given focus to help researchers achieve improved efficiency. However, reprogramming is a dynamic process and needs several weeks to complete. Moreover, the existing protocols are non-standardized and therefore, vary with different laboratories and/or researchers.
Irrespective of the reprogramming method, a significant amount time has been spent in the laboratory investigating sensitive cultures, monitoring stem cell colony emergence in non-physiological environments, assessing colony quality based on morphological markers, and tracking and maintaining established iPSC clones.
The IncuCyte® Live Content Imaging system offers an integrated time-lapse, whole well imaging acquisition and analysis package for facilitating iPSC work flow by offering monitoring and characterization tools for stem cell colonies and rare events. The software tools in this package not only simplify the entire well scanning but also facilitate clone selection, clone tracking, and culture maintenance. These tools:
- Simplify observation of colonies back in time, allowing scientists to identify when they have emerged and investigate what they resembled along the way
- Shorten the time spent by cultures outside the incubator
- Can be employed to standardize protocols
- Reduce the time spent by researchers on stereoscopes and in culture hoods (Figure 1)
This article discusses the use of Life Technologies CytoTune®-IPS Sendai Reprogramming Kit to demonstrate these software and hardware tools by delivering the Yamanaka factors to both human neonatal and adult fibroblasts for capturing successful reprogramming in both feeder and feeder free conditions (Figure 2).
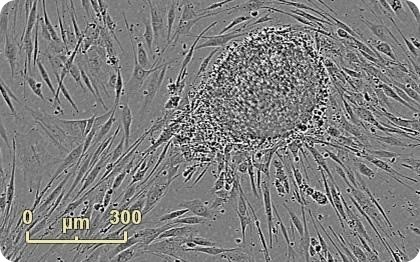
Figure 1. Emerging colony during fibroblast reprogramming. Human neonatal fibroblasts were reprogrammed using Life Technologies CytoTune®-IPS Sendai Reprogramming Kit. An emerging colony grown on mouse embryonic fibroblast feeder cells is observed 22 days into the reprogramming process. Images were acquired at 4x in Whole Well Imaging Mode.


Figure 2. Schematic overview of the CytoTune® reprogramming procedure for fibroblasts, from Life Technologies.
Approach and methods
Cell culture
Cells were grown to 80% to 90% confluent at the time of adding CytoTune®-IPS Sendai Reprogramming reagent in order to achieve optimized reprogramming. This study involved the initiation of human neonatal foreskin fibroblasts and human adult dermal fibroblasts into culture and their growth in T75 cell culture flasks to 80% confluence as determined by an IncuCyte®.
DMEM (Gibco) supplemented with 10% ES qualified fetal bovine serum (Gibco) was used to culture both fibroblast lines. The passage number is kept low by passing each fibroblast line one time because the lines are primary sourced cell lines.
Reprogramming procedure
Before initiating the reprogramming experiment, TrypLE™ Express (Gibco) was used to lift the fibroblasts and each cell line was seeded into two wells of a 6-well plate (Corning) at 2×105 cells per well.
The plates were then transferred into IncuCyte® equipped with a 4x objective, and incubated in a HeraCell cell culture incubator at 37˚C and 6% CO2. Confluence was monitored by collecting whole well images in phase-contrast every 4 hours.
After 48 hours, the cells were grown to 80% to 90% confluent and then the reprogramming experiment was started. The reprogramming event was performed by carefully following Life Technologies’ CytoTune®-IPS Sendai Reprogramming protocol.
The schematic overview of the CytoTune®-IPS Sendai protocol for reprogramming is shown in Figure 2. On Day 0, cMYC, OCT4, SOX2, and KLF4 consisting of Sendai Viruses were included separately to the cultures.
On Day 1, the media was removed and replaced with fresh fibroblast medium. Phase contrast images were then used to qualitatively monitor cell health (Figure 3).
Alternatively, the amount of toxicity at the time of reprogramming reagent addition can be more quantitatively evaluated by adding a membrane impermeable DNA dye (e.g. YOYO-®1 (Life Technologies), CellTox Green™ (Promega)) to a test well.
On Day 7, transduced cells were collected, counted, and then plated at 50,000 cells per well into 6-well plates comprising of either a coating of Geltrex™ (Life Technologies) or a feeder layer of Mouse Embryonic Fibroblasts (MEFs; Millipore).
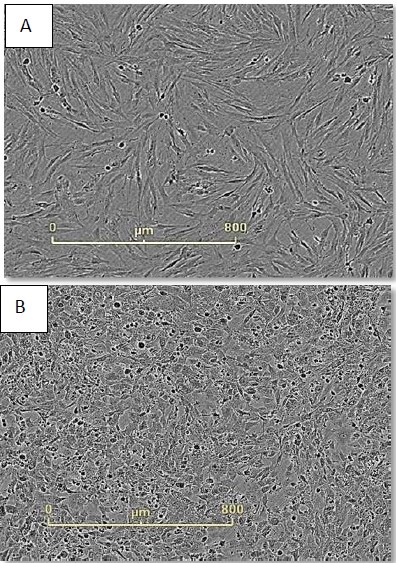
Figure 3. Monitor cell health during reprogramming. Neonatal foreskin fibroblasts imaged just before (A) and 48 hours after (B) CytoTune® addition. Cell morphology has changed dramatically during this time frame and continues to do so during the first seven days of reprogramming.
On Day 8, the media was replaced with either Essential 6™ (Gibco) media supplemented with bFGF in the feeder free culture or Knockout™ Serum Replacement consisting of DMEM-F12 media (Gibco) in the MEF feeder system. Media replacement was carried out daily thereafter.
Wells were imaged using the “whole well” scan mode every 4 hours in order to examine the entire surface area in the IncuCyte® software. The automated navigation and panning tools were used to visually inspect each well daily.
The emergence of colonies was observed at approximately Day 12, i.e., 5 days after reseeding (Figure 4). Colonies were labeled and tracked using software Point of Interest (POI) tools as soon as they were detected as possible pluripotent colonies (Figure 5).
Together, time-lapse imaging and integrated software tools allow studying the complete timeline of the emergence and evolution of the colonies, from reseeding to colony picking.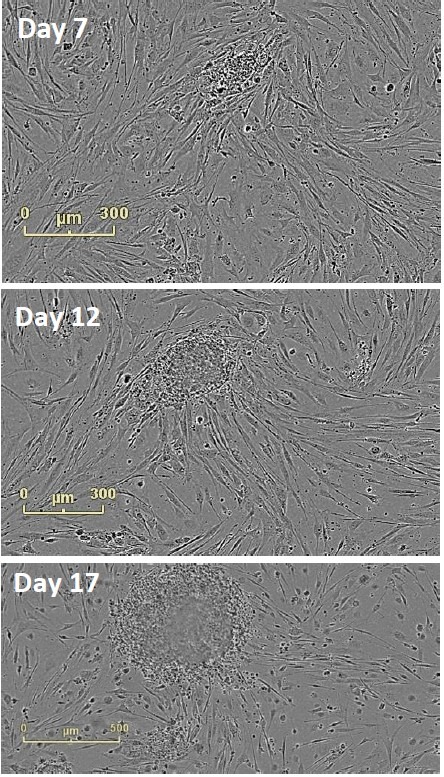
Figure 4. Follow Colony Emergence. An emerging colony at day 7, day 12, and day 17. This iPSC colony was derived from neonatal fibroblasts, grown on mouse embryonic fibroblasts, and identified at day 12. Moving backwards through the images, colonies can be examined on the day they are passaged onto feeder cells.
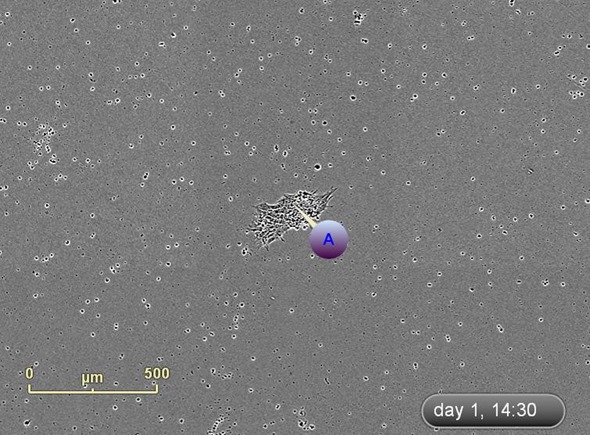
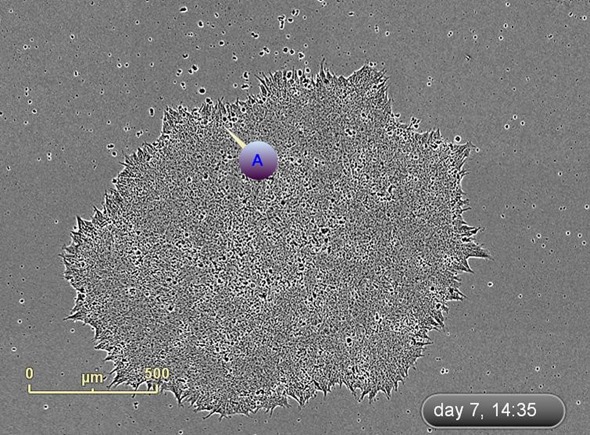
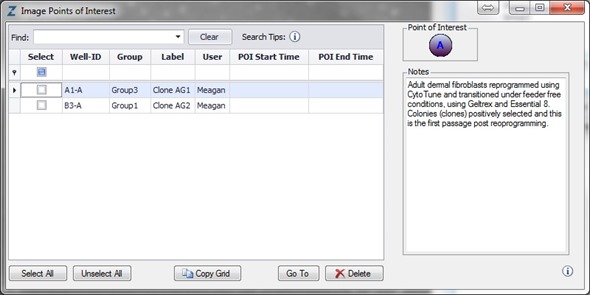
Figure 5. Label and track colonies. Induced pluripotent colony derived from Human adult dermal fibroblasts and cultured feeder free, on Geltrex™. Colony was tagged as a point of interest and easily found and followed on a daily basis. Top image represents a colony identified early in the process and labeled as a point of interest. The bottom image shows the same colony six days later. The point of interest manager is used to easily navigate between different colonies, take notes on colony characteristics, assign beginning (day of seeding) and end (day of picking) points to each colony, and label colonies appropriately.
Initial passaging and subsequent maintenance
Round, tight border, and several small cells with higher nuclear to cytoplasmic ratio are the morphological characteristics of pluripotency typical of stem cell colonies.
These characteristics can be normally seen at >Day 16 after the completion of reprogramming. Colonies for selection and passaging were identified using these characteristics.
Here, colonies were picked between Days 19-22 by first applying a digital mark using IncuCyte® software tools around the colonies chosen for picking and expansion within a whole well image (Figure 6).
The digital marks in the software were then transferred into physical markings on the actual plate of cells using the IncuCyte® Marking Tool (Figure 6).
The culture plates were then placed in the tissue culture hood, followed by micro dissecting the colonies into several smaller pieces using a custom picking tool. Each colony was labeled based on the colony from which it was dissected and plated into 6-well plates prepared with MEF feeders (feeder system) or Geltrex™ coating (feeder free system), in KSR™ or complete Essential 8™ media, respectively.
At this time, Essential 8™ was used in place of Essential 6™ + bFGF. Phase-contrast images were captured every 4 hours. It has been demonstrated that if colonies are not pluripotent during the time of selection, morphological changes related to differentiation can be clearly observed within the first several passages after selection.
The use of the IncuCyte® simplifies the tracking of the transformation of colonies into culture after positive selection.
Reprogramming efficiency calculations
Alkaline phosphatase activity is closely associated with pluripotency and the standard method for determining reprogramming efficiency is terminal staining for alkaline phosphatase activity.
Here, a red fluorescent/colorimetric alkaline phosphatase reagent (Vector Laboratories; Vector® Red Alkaline Phosphatase Substrate Kit 1) was used to stain a duplicate well of reprogrammed cells (neonatal fibroblasts grown on a MEF feeder layer) and the integrated image analysis tools featured in the IncuCyte® software package were used to count the colonies (Figure 7).
The minimum size of colonies counted was defined by applying a size filter of 5×104 μm2. The total number of colonies counted from a starting density of 5×104 cells that were replated at Day 7 subsequent to the addition of Yamanaka factor was 147, resulting in a final reprogramming efficiency of 0.294%.
Post-reprogramming quality control and pluripotency validation
For routine maintenance of the cell culture, the whole well scans and software panning/navigation tools in the IncuCyte® were used to monitor picked colonies for evidence of differentiation.
Preparations were initiated for colony passage based on confluence metrics (feeder free), colony diameter measurements, and observations of colony spacing. Decisions were also made to clean areas of differentiation or colony removal.
Plates were appropriately labeled using the IncuCyte® Marking Tool (Figure 6), thus providing a point of reference for further treatment in a tissue culture hood at a later stage. After cleaning and removal, 70% ethanol was used to wipe the marks from the plate and scanning was restarted by placing the vessel back into the IncuCyte®.
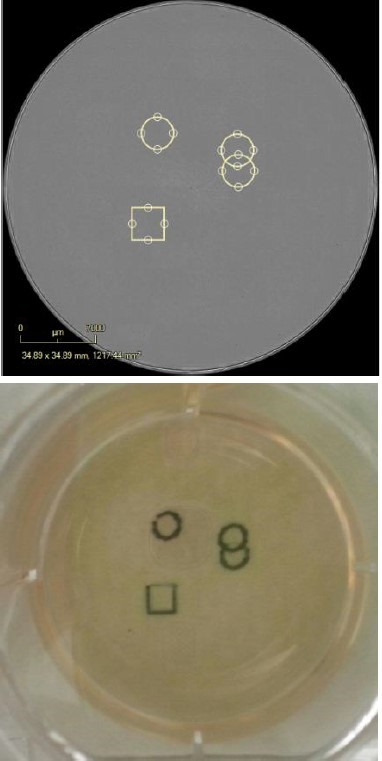
Figure 6. Digitally and physically mark the vessel. Digitally place pen marks around the colonies for selection. Place the Marking Tool in the objective location and the pen will place physical marks as defined in the software. The vessel can then be removed from, taken into the biosafety cabinet and the colonies of interest can be removed for expansion.
Pluripotency was confirmed by staining the clones for the surface marker SSEA4. The SSEA4 is a pluripotency indicator for human embryonic stem cells and an indicator intracellularly for the existence of OCT4 protein, which is highly expressed in pluripotent stem cells.
It is expressed during early development. Identical protocols were used to perform single (SSEA4) and double (SSEA4 + OCT4) label immunocytochemistry. After fixing the cells with 4% paraformaldehyde, they were labeled with mouse anti-human SSEA4 (Life Technologies) alone (Figure 7) or along with rabbit anti-human OCT4 (Life Technologies) (Figure 8).
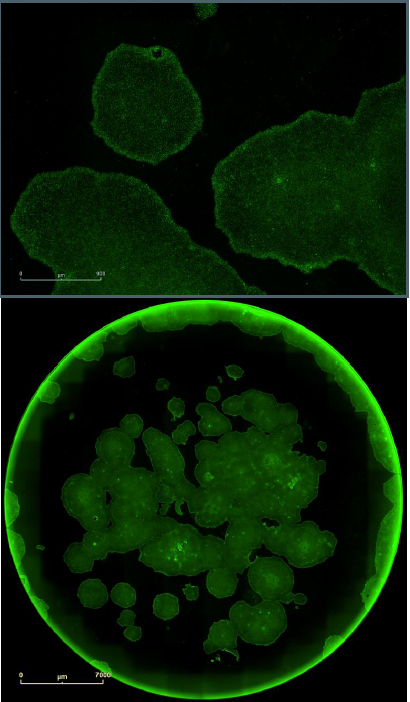
Figure 7. Single label confirmation of pluripotency. SSEA4 primary antibody paired with Alexa Fluor® 488 secondary antibody confirmed pluripotency on a clone derived from foreskin fibroblasts and grown in feeder free conditions on Geltrex™. Upper images provides a magnified view of the colonies with the cell surface labeling, while the lower images provides the whole well overview in green fluorescence.
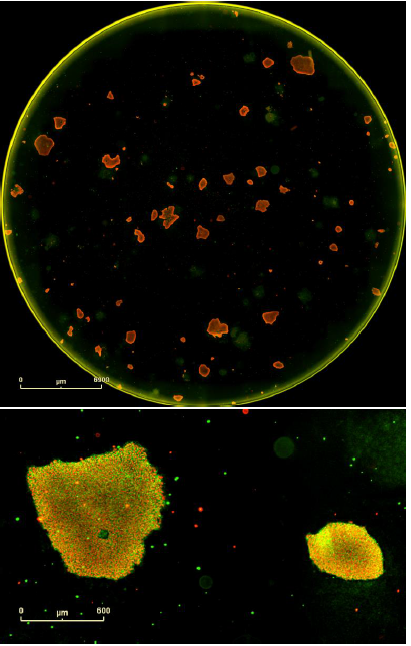
Figure 8. Double label confirmation of pluripotency. SSEA4 primary antibody paired + Alexa Fluor® 488 secondary antibody combined with OCT4 primary antibody + Alexa Fluor® 594 secondary antibody. Top image is the whole well overview and the bottom image is a magnified image of two colonies from this well expressing both markers of pluripotency. These are iPSCs derived from human adult dermal fibroblasts and grown on mouse embryonic fibroblast feeder cells.
Both single and double staining protocols indicated that SSEA4 is expressed by these clones. The double staining protocol also revealed that OCT4 is expressed by this clone. These results confirm the successful reprogramming to a pluripotent state.
Key features of CellPlayer™ kinetic stem cell application
The following are the key features of CellPlayer™ kinetic stem cell application:
- Stem cell colonies can be imaged by moving forward and backward through time: The kinetic aspect of the imaging system of the IncuCyte® enables researchers to detect colony emergence at the earliest point and then follow backward to see how they appeared at the initial stage.
- Stable environment for sensitive cells: Sensitive cultures can be scanned and investigated while the cells are within the incubator owing to the transfer of a considerable amount of the workflow to the IncuCyte®.
- IncuCyte® Marking Tool: The IncuCyte® Marking Tool transferring digital marks made within the software directly to actual plates. The vessel can then be transferred from the IncuCyte® to the biosafety cabinet and stereoscope for further manipulation.
- Software features facilitate recordkeeping and standardizing protocols: Software features allow users to easily scan through images, note down key associated characteristics, label points of interest, and collect quantitative data from images with integrated image processing tools.
- Facilitates training and collaboration: Since the generation and organization of images, movies, and image collections is simplified, the knowledge can be easily shared between collaborators and colleagues and training can be efficiently provided to researchers new to the field.
- Less congestion on shared equipment: Stereo microscopes and tissue culture hoods are shared in the laboratory. Congestion can be alleviated by integrating the IncuCyte® into the work flow.
Acknowledgements
Produced from materials originally authored by Meagan A. Roddy1, Rene H. Quintanilla Jr.2, Patricia M. Garay1, Uma Lakshmipathy2, and Daniel M. Appledorn1 from:
1Sartorius – Ann Arbor, MI
2Life Technologies – Carlsbad, CA
References
- Takahashi, K. and Yamanaka, S. (2006) Cell 126, 663-676
- Takahashi, K et al (2007) Cell 131, 861-872
- Stadfeld, M. et al (2008) Science 322, 945-949
- Carey, B.W. et al (2008) PNAS 106, 157-162
- Maherali, N. and Hochedlinger, K. (2008) Cell Stem Cell 3, 595-605
- Fusaki, N. et al (2009) Proc. Jpn. Acad. Ser. 85, 348-362
- O’Connor, M.D. et al (2008) Stem Cells 26,1109-1116
 Sartorius
Sartorius
Sartorius is a leading international pharmaceutical and laboratory equipment supplier. With our innovative products and services, we are helping our customers across the entire globe to implement their complex and quality-critical biomanufacturing and laboratory processes reliably and economically.
The Group companies are united under the roof of Sartorius AG, which is listed on the Frankfurt Stock Exchange and holds the majority stake in Sartorius Stedim Biotech S.A. Quoted on the Paris Stock Exchange, this subgroup is comprised mainly of the Bioprocess Solutions Division.
Innovative Technologies Enable Medical Progress
A growing number of medications are biopharmaceuticals. These are produced using living cells in complex, lengthy and expensive procedures. The Bioprocess Solutions Division provides the essential products and technologies to accomplish this.
In fact, Sartorius has been pioneering and setting the standards for single-use products that are currently used throughout all biopharmaceutical manufacturing processes.
Making Lab Life Easier
Lab work is complex and demanding: Despite repetitive analytical routines, lab staff must perform each step in a highly concentrated and careful way for accurate results.
The Lab Products and Services Division helps lab personnel excel because its products, such as laboratory balances, pipettes and lab consumables, minimize human error, simplify workflows and reduce physical workloads
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.