After the rapid incline in COVID-19 cases and deaths in India over recent weeks, travel bans and restrictions to and from India have been imposed by at least 20 countries. Once again, the world has arrived at a crossroads of viral evolution.
When reports first came in regarding the Indian variants, scientists were notified that two potent mutations were simultaneously occurring on the viral spike protein, namely E484Q and L452R5, both demonstrating higher infectivity and immune evasion.
Thanks to the rapid sequence collection from all over India, the analytical progress of and analysis has led to an increase in critically understanding the combined effect of these two mutations, as well as other mutations fostered by the variants, designated B.1.617 by PANGOLIN classification.
Predicated on a phylogenetic study by Indian from the retrieved sequences Council of Medical Research (ICMR)6, in total, 23 non-synonymous changes at the spike protein were distinguished; of which seven frequently found signature mutations were identified (G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H).
The study further categorized the B.1.617 lineages into three sub-clusters, which vary in their exact mutations. Notably, T95I was shared by a clade of sequences; second H1101D and third V382L, V1175Y (Figure 1.).
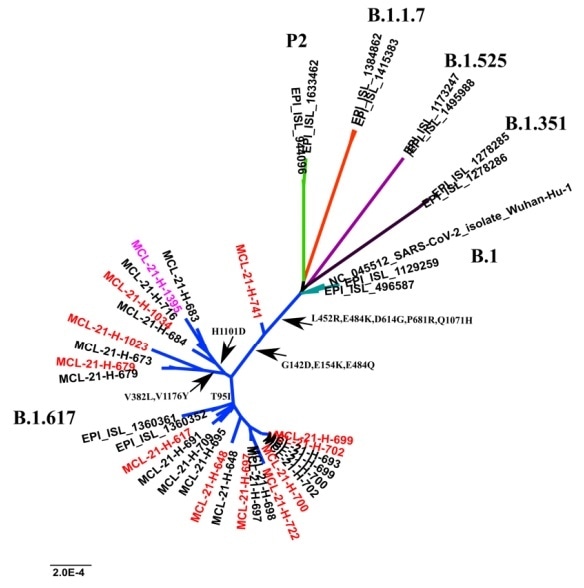
Figure 1. A phylogenetic tree of representative SARS-CoV-2 genomes and the occurring mutations in the spike protein in the sub-clusters pf lineage B.1.617. Image Credit: ACROBiosystems
Researchers offered multiple explanations for the devastating spread of B.1.617 in India:
1) Both L452R and E484Q have an increased stabilizing effect on the spike protein that could facilitate more effective binding to the human ACE2 receptor, resulting in an increase in host cell entry4.
2) P681R in the S1/S2 furin cleavage site may allow more severe proteolytic activation of the spike protein, leading to enhanced infectivity2.
3) The new combinations of mutations possess better immune evasion, e.g., reduced binding by monoclonal antibodies (mAbs) and incompatibility of current vaccines3.
By neutralizing tests with convalescent or vaccinated sera against the mutated spike protein, the augmented fitness of B.1.617 was verified. In a recent study issued in the pre-print repository medRxiv3, plasma from COVID-19 convalescent patients show a decrease in neutralization against the variant when compared against the Wuhan WT spike protein.
However, the reduction in the capacity to neutralize was reduced to around 2-fold and not as potent as B.1.351 (~6-fold reduction) (Figure 2A.).
In the neutralization test with serum from Comirnaty/BNT162b2 (Pfizer–BioNTech vaccine) individuals that had received the vaccine, the inhibition of viral entry mediated by B.1.617 spike protein was observed to be inefficient but also less pronounced (~3-fold reduction) when compared to B.1.351 (>11-fold reduction) (Figure 2B.).
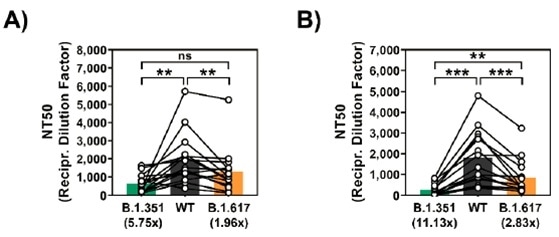
Figure 2. (2A) Diminished neutralization by plasma from COVID-19 convalescent patients against SARS-CoV-2 WT, B.1.351, and B.1.617. (2B) Diminished neutralization by plasma from Comirnaty/BNT162b2 vaccinated patients against SARS-CoV-2 WT, B.1.351, and B.1.617. Image Credit: ACROBiosystems
Results acquired by ICMR discovered that recipients of Bharat Biotech's BBV152 (Covaxin) possessed the ability to neutralize VUI B.1.617, albeit with a slightly lower efficacy6.
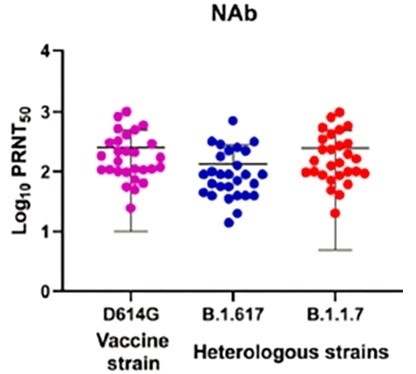
Figure 3. Neutralizing response of the individual sera (n=28) vaccinated with BBV152 (Covaxin) collected during phase II clinical trial for the prototype B1 (D614G) (pink), B.1.1.7 (red), B.1.617 (blue). Image Credit: ACROBiosystems
These findings produced in these preliminary studies produce are appropriate insofar as they may help avoid vast spread and transmission of the variant.
In order to introduce the requisite future public health interventions, genomic surveillance will be necessary on a continuous basis to keep track of the mutations on B.1.617 and other potential variants of SARS-CoV-2.
Consequently, ACROBiosystems has sped up the development of B.1.617 recombinant antigens to facilitate a better understanding of the transmissibility and infectivity of the variant.
References
- Nextstrain / ncov / global
- Cherian, S., Potdar, V., Jadhav, S., et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv 2021.04.22.440932; doi: https://doi.org/10.1101/2021.04.22.440932
- Hoffmann, M., Hofmann-Winkler, H., Krüger, N., et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv 2021.05.04.442663; doi: https://doi.org/10.1101/2021.05.04.442663
- Kumar, V., Singh, J., Hasnain, S. E., Sundar, D. Possible link between higher transmissibility of B.1.617 and B.1.1.7 variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. bioRxiv 2021.04.29.441933; doi: https://doi.org/10.1101/2021.04.29.441933
- Ranjan, P., Neha, Devi, C., Das, P., et al. Bioinformatics analysis of SARS-CoV-2 RBD mutant variants and insights into antibody and ACE2 receptor binding. bioRxiv 2021.04.03.438113; doi: https://doi.org/10.1101/2021.04.03.438113
- Yadav, P. D., Sapkal, G. N., Abraham, P., et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. bioRxiv 2021.04.23.441101; doi: https://doi.org/10.1101/2021.04.23.441101
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.