Like its predecessor, SARS-CoV-2 mainly targets ACE2 receptors for viral entry and infection. Biologically, ACE2 works to counteract the renin-angiotensin system in order to convert Angiotensin peptides which are hormones regulating vasoconstriction and vasodilation in cells.
The ACE2 receptor can be found in high levels in the heart, kidneys, and testis. ACE2 structures are made up of the functional extracellular peptidase domain, followed by the collectrin-like domain or the neck domain. As seen in Figure 10A, in these structures, the transmembrane domain and 40 residue intracellular segments are not resolved.
The secretion of active ACE2 peptidase is enabled by a protease cleavage site in the neck domain ACE2. As seen in Figure 10A, ACE2 dimerizes mediated primarily by the neck domain via H-bond and cation- interaction into an open and closed conformation.13
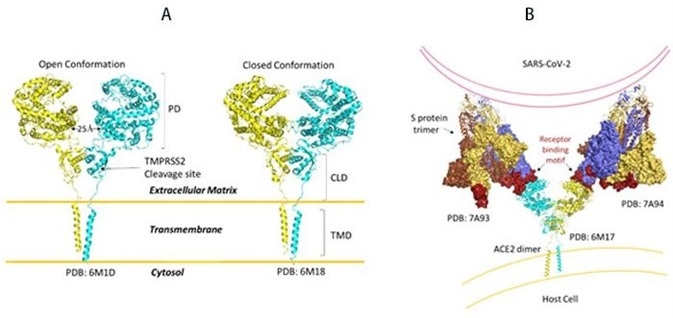
Figure 10. A) Full-length structure of ACE2 in open and closed homodimer states. B) Modeled interaction of S trimer to ACE2 dimer during viral infection. Image Credit: ACROBiosystems
It remains to be seen whether clustering of S trimer and ACE2 dimers occurs and enables the fusion of entry of SARS-CoV-2.
The open conformation of ACE2 binds to the RBD domain of S protein. As seen in Figure 10B, docking S protein trimer shows that each ACE2 dimer can bind to two S protein trimers simultaneously.13
The full-length structure of ACE2 in open and closed homodimer states is shown in Figure 10A, and the modeled interaction of S trimer to ACE2 dimer during viral infection is demonstrated in Figure 10B. It is not yet known whether clustering of S trimer and ACE2 dimers happens to allow fusion of the entry of SARS-CoV-2.
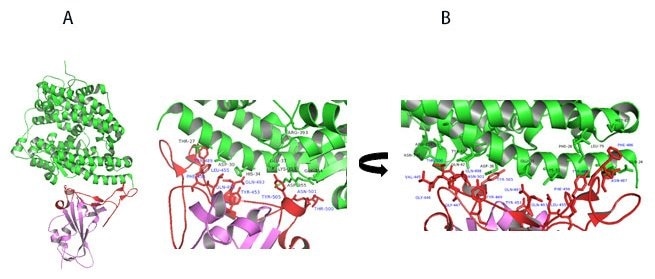
Figure 11. A) Co-crystal of RBD (purple) bound to ACE2(green). B) Polar interaction within the RBM that enables tight binding to ACE2. RBM is colored red. PDB: 6M0J. Image Credit: ACROBiosystems
Upon further study of residues involved in RBD-ACE2 interaction, the receptor-binding motif (RBM) forms physical contact with ACE2 receptors. As seen in Figure 11A, key residues on the RBM within the RBD make physical contact with ACE2 receptors and result in a large buried surface of 1,687 Å2 created by interactions of 17 RBD residues to 20 ACE2 residues.
Similar to SARS-CoV, key interactions are mediated by polar residues (13 H-bond and 2 salt bridges).1, 14 An extended loop region of RBD which contains α4, α5 helices and loops and β5, β6 strands form a bridge with the α1 helix of ACE214 as shown in Figure 11A-B.
Receptors for featured super stable S protein trimer from ACROBiosystems
ACROBiosystems provides ACE2 proteins with numerous species and tags to facilitate in-depth study of S-Protein/ACE2 binding mechanism and examine the efficacy of therapeutics and vaccines.
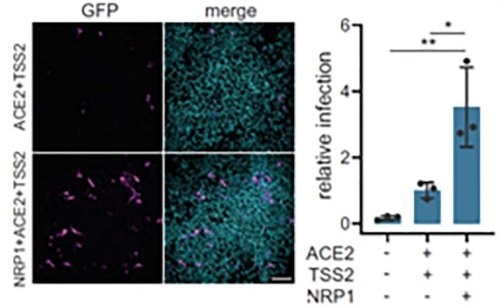
Figure 12. Neuropilin-1 and ACE2 co-expression dramatically improve viral entry. Image Credit: ACROBiosystems
Further to ACE2, Neuropilin-1 receptor, which binds to furin cleaved substrates, has demonstrated that it facilitates SARS-CoV-2 viral entry, especially in the nasal cavity cells. Viral entry was significantly improved by expression of both Neuropilin-1 and ACE2 receptors in HEK-293T cells instead of ACE2 alone, as seen in Figure 12.15
Neuropilin-1 receptor is thought to promote entry of SARS-CoV-2 due to the low level of expression of ACE2 receptor in respiratory and epithelial cells, enabling cell, vascular, and tissue penetration. ACROBiosystems provides their tagged and biotinylated Neuropilin-1 with high purity and bioactivity.
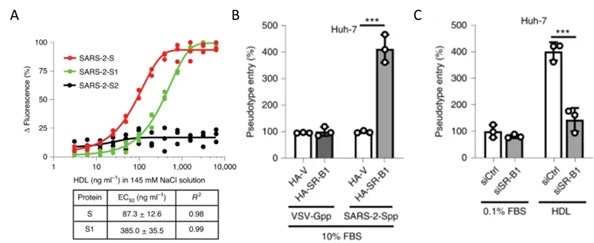
Figure 13. A) S protein S1 subunit binds to components of HDL. b) HDL receptor SR-B1 enhances SARS-CoV-2 entry. Image Credit: ACROBiosystems
As shown in Figure 13A, recently, the S1 domain of SARS-CoV-2 has been shown to bind to Cholesterol and components of high-density lipoprotein (HDL)16. So, the HDL scavenger receptor B type 1 (SR-B1) has been involved in enhanced ACE2 dependent viral entry as demonstrated in the pseudo viral entry assay.
As seen in Figures 13B-C, the expression of SR-B1 improves viral entry while knockdown of SR-B1 worsens viral entry.16 SR-B1 acts as a host factor promoting SARS-CoV-2 entry and is co-expressed with ACE2 in pulmonary tissue.
Large-scale screening of 5054 human membrane proteins recently identified a panel of receptors that have a strong affinity to SARS-CoV-2 spike protein and could lead to alternative viral entry pathways which could explain disparate symptoms of COVID-19.17
Other recent studies have found that other receptors are also involved in SARS-CoV-2 infection, such as restriction enzymes and proteases.18 According to the reference description, ACROBiosystems attempted to verify binding affinity between SARS-CoV-2 S protein with the human receptors below:
Table 2. Kd measurement of the interaction of SARS-CoV-2 S protein with human receptors. Source: ACROBiosystems
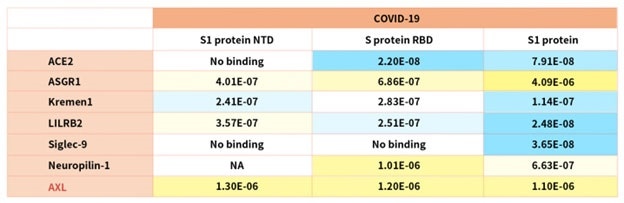
Notably, the latest research demonstrated that the tyrosine-protein kinase receptor UFO (AXL) specifically might interact with the SARS-CoV-2 Spike protein and take a key role in promoting viral infection of the human respiratory system.
References
- B, H.; H, G.; P, Z.; ZL, S., Characteristics of SARS-CoV-2 and COVID-19. Nature reviews. Microbiology 2020.
- E, P.; M, K.; U, G.; DH, H.; N, P.; F, C.; M, S.; S, A. K.; L, S., Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. The Lancet. Infectious diseases 2020, 20 (9).
- Z, Z.; X, L.; X, S.; W, W.; GA, M.; Y, Z., From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 2020, 21 (1).
- JM, W.; SE, D.; M, B.; K, S., Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Critical reviews in biochemistry and molecular biology 2008, 43 (3).
- T, T.; M, B.; JA, J.; GR, W.; S, D., Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research 2020, 178.
- Buchholz, U.; Bukreyev, A.; Yang, L.; Lamirande, E.; Murphy, B.; Subbarao, K.; Collins, P., Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proceedings of the National Academy of Sciences of the United States of America 2004, 101 (26).
- Traggia, i. E.; Becker, S.; Subbarao, K.; Kolesnikova, L.; Uematsu, Y.; Gismondo, M.; Murphy, B.; Rappuoli, R.; Lanzavecchia, A., An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature medicine 2004, 10 (8).
- J, S.; Y, W.; C, L.; G, Y.; Q, G.; A, A.; F, L., Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America 2020, 117 (21).
- DJ, B.; AG, W.; P, X.; C, R.; SR, M.; PB, R.; JJ, S.; SJ, G., Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588 (7837).
- R, H.; RJ, E.; K, M.; K, J.; V, S.; SMC, G.; M, K.; D, L.; R, P.; AL, H.; MJ, B.; BF, H.; P, A., Controlling the SARS-CoV-2 spike glycoprotein conformation. Nature structural & molecular biology 2020, 27 (10).
- CL, H.; JA, G.; JM, S.; AM, D.; HC, K.; K, J.; KC, L.; D, W.; AG, L.; Y, L.; CW, C.; PO, B.; CK, H.; NV, J.; J, L.-M.; AW, N.; J, P.; N, W.; D, A.; JJ, L.; GC, I.; JA, M.; IJ, F.; JS, M., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science (New York, N.Y.) 2020, 369 (6510).
- D, W.; N, W.; KS, C.; JA, G.; CL, H.; O, A.; BS, G.; JS, M., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020, 367 (6483).
- R, Y.; Y, Z.; Y, L.; L, X.; Y, G.; Q, Z., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 2020, 367 (6485).
- J, L.; J, G.; J, Y.; S, S.; H, Z.; S, F.; Q, Z.; X, S.; Q, W.; L, Z.; X, W., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581 (7807).
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L. D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Meer, F. v. d.; Kallio, K.; Kaya, T.; Anastasina, M.; Smura, T.; Levanov, L.; Szirovicza, L.; Tobi, A.; Kallio-Kokko, H.; Österlund, P.; Joensuu, M.; Meunier, F. A.; Butcher, S. J.; Winkler, M. S.; Mollenhauer, B.; Helenius, A.; Gokce, O.; Teesalu, T.; Hepojoki, J.; Vapalahti, O.; Stadelmann, C.; Balistreri, G.; Simons, M., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370 (6518), 856-860.
- C, W.; L, W.; Q, Y.; X, W.; J, Z.; X, Y.; Y, Z.; C, F.; D, L.; Y, D.; J, S.; J, G.; X, Y.; Y, W.; X, W.; J, L.; H, Y.; H, L.; Z, Z.; R, W.; P, D.; Y, Z.; F, Y.; W, Z.; N, W.; Y, P.; H, L.; J, F.; C, Q.; W, C.; Q, G.; R, Z.; Y, C.; H, Z., HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nature metabolism 2020, 2 (12).
- Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; Zhang, J.; Shen, G.; Jiang, X.; Yang, J.; Zheng, X.; Xu, J.; Zhang, C. C.; Lan, F.; Qu, D.; Zhao, Y.; Xu, G.; Xie, Y.; Luo, M.; Lu, Z., Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv 2020.
- Wang, S., Qiu, Z., Hou, Y. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31, 126–140 (2021).
- L, Z.; CB, J.; H, M.; A, O.; H, P.; BD, Q.; ES, R.; A, P.; A, V.; MS, S.; W, L.; T, I.; C, R.; M, F.; H, C., SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nature communications 2020, 11 (1).
- L, Y.; X, W.; KE, P.; C, T.-T.; TP, N.; Y, W.; A, B.; WE, D.; A, D.; C, C.; K, V.; SB, E.; SF, S.; JE, L.; JB, M.; A, R.; A, B.; PC, S.; CA, K.; NV, D.; K, S.; J, L., Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183 (3).
- Zhang, W.; Davis, B. D.; Chen, S. S.; Martinez, J. M. S.; Plummer, J. T.; Vail, E., Emergence of a novel SARS-CoV-2 strain in Southern California, USA. Preprint 2021.
- Wang, P.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P. D.; Graham, B. S.; Mascola, J. R.; Chang, J. Y.; Yin, M. T.; Sobieszczyk, M.; Kyratsous, C. A.; Shapiro, L.; Sheng, Z.; Nair, M. S.; Huang, Y.; Ho, D. D., Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv 2021.
- PANGO lineages. https://cov-lineages.org/global_report.html.
- Wu, K.; Werner, A. P.; Moliva, J. I.; Koch, M.; Choi, A.; Stewart-Jones, G. B. E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B. S.; Carfi, A.; Corbett, K. S.; Seder, R. A.; Edwards, D. K., mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021.
- Xie, X.; Zou, J.; Fontes-Garfias, C. R.; Xia, H.; Swanson, K. A.; Cutler, M.; Cooper, D.; Menachery, V. D.; Weaver, S.; Dormitzer, P. R.; Shi, P.-Y., Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021.
- Novavax, Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. 2021.
- Greaney, A. J.; Loes, A. N.; Crawford, K. H. D.; Starr, T. N.; Malone, K. D.; Chu, H. Y.; Bloom, J. D., Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. 2021.
- Luan, B.; Wang, H.; Huynh, T., Molecular Mechanism of the N501Y Mutation for Enhanced Binding between SARS-CoV-2’s Spike Protein and Human ACE2 Receptor. bioRxiv 2021.
- RL, G.; A, N.; P, C.; J, B.; B, H.; J, M.; G, H.; J, C.; B, M.; V, S.; I, S.; P, K.; AC, A.; J, V. N.; KL, C.; M, D.; G, O.; AE, S.; TR, H.; PJ, E.; RE, H.; NL, K.; J, S.; DR, P.; P, K.; L, S.; DM, S., Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021.
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.