The S protein of SARS-CoV had been studied extensively before the SARS-CoV-2 outbreak. Among other structural proteins of SARS-CoV-2, the S protein was discovered to be highly immunogenic and effective at neutralizing antibodies recovered from SARS patients were directed at S protein.6,7
Research into therapeutics and vaccines against SARS-CoV-2 was focused immediately on the S protein. Although the four structural proteins of the novel SARS-CoV share over 90% in sequence identity with SARS-CoV-2, S protein consisting of S1 and S2 subunits diverged had just 77% sequence similarity.1
The S1 subunit contains the NTD or the N-terminal domain and Receptor binding domain (RBD), which binds to the human angiotensin-converting enzyme-2 (ACE2) mainly through direct interaction with the receptor-binding motif (RBM).
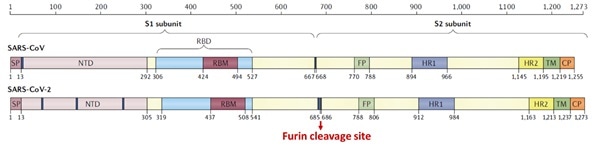
Figure 5. Structural schematic of S protein between SARS-CoV and SARS-CoV-21. Image Credit: ACROBiosystems
The S2 subunit is made up of the heptad repeats, fusion peptide, transmembrane region, and cytoplasmic domain, as seen in Figure 5. Compared with SARS-CoV, the high transmissibility of SARS-CoV-2 has been posited to happen.
This is due to the insertion of a polybasic cleavage site between S1 and S2 subunits which decreases the stability of S protein and facilitates conformational alteration needed to bind the ACE2 receptor.1
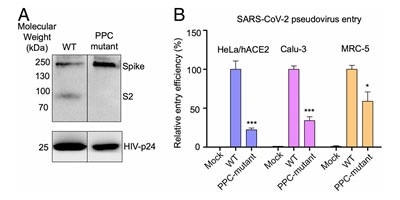
Figure 6. S protein cleavage occurs and necessary for viral infection. A) Mutation of furin (PPC) site abolishes cleavage. B) PPC mutant have lower viral entry efficiency compared to WT. Image Credit: ACROBiosystems
Lentivirus presenting S protein in a pseudo viral entry assay actually demonstrates that protease cleavage of S protein happens during viral entry.8 Figure 6A demonstrates that mutating the putative polybasic cleavage site on S protein abrogated the cleavage.
Viral entry efficiency in ACE2 expressing lung cancer cell lines and HeLa cells was lowered significantly in cleavage site mutants of S protein8, as demonstrated in Figure 6B. The necessity of cleavage of S protein for viral entry is confirmed by these results.
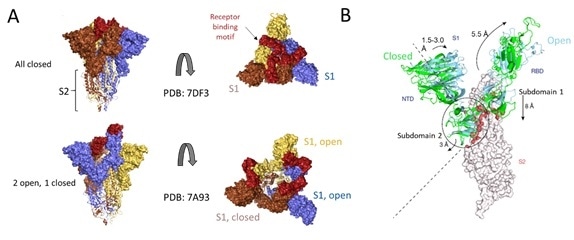
Figure 7. Cryo-EM structure of S trimer. A) Depiction of S1 subunits in open and closed RBD conformation as surface representation and S2 subunits in ribbon form. B) Changes in S1 subunit post ACE2 receptor binding. Image Credit: ACROBiosystems
Cryo-EM structures of S protein have accelerated further understanding of the structural underpinnings of S protein function in viral attachment and entry. Structural analysis of the S protein showed a trimeric structure with a central stalk consisting of 3 S2 subunits, as seen in Figure 7A, which is topped by 3 S1 subunits.9
The S1 subunit is dynamic, with conformation changes oscillating between closed and open states. The RBD is accessible to binding via the ACE2 receptor in the S1 open conformation.
These rearrangements and movements by RBD and NTD domains of the S1 subunit are driven by the energetics of ACE2 binding.9 ACE2 binding modifies the open RBD conformation by a rigid-body rotation of the domain, moving it further 5.5 Å away from the trimer axis while the NTD domain moves 1.5 to 3 Å9.
The NTD-associated subdomain two moves by 3 Angstrom while the RBD-associated subdomain 1 moves about 8 Å while, as shown in Figure 7B.
Via a series of stacking in the closed conformation, the NTD associated subdomain 2 interacts with a short helix and loop of the S2 subunit, as seen in Figure 7B. In the open conformation, these interactions are lost, leaving a channel in S2, which is disordered and exposed for the next step of membrane fusion.9
The energetics of ACE2 binding events fuel the major rearrangement of the S trimer. ACE2 binding favors RBD open conformation, and sequential opening and binding of RBD in the S trimer results in a fully open state and a notable decrease in contact between S1 neighbors and S2 trimeric core.
The fully open state leaves a 65 Å deep, 50 Å diameter around the trimer axis9. This new open structure also results in solvent exposure of the fusion peptide on S2, in addition to the major helical rearrangement resulting in host cell fusion.
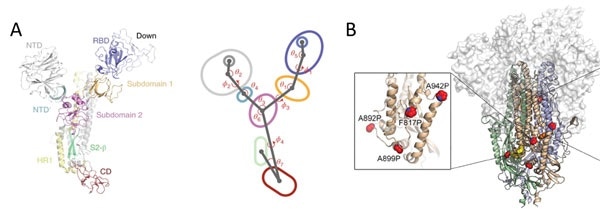
Figure 8. A) S protein is a highly dynamic structure with multiple centroid rotating along with various angles and dihedrals10. B) 6 Proline mutations in the S protein timer stabilizes the prefusion state. Image Credit: ACROBiosystems
Domains in the S1 subunit and S2 subunit are made up of centroids with different angles and dihedrals due to the dynamic nature of S protein,10 as seen in Figure 8A. This means mutations at sites intersecting these centroids will influence the conformation landscape of the S protein and affect therapeutic and vaccine development.
Prefusion stable S proteins are also required to grow recombinant protein expression yield with homogeneous preparations.11, 12 As seen in Figure 8B, proline substitutions (2P and 6P) have been identified to stabilize loops and cap helices in the prefusion state in addition to the introduction of disulfide bonds prevents conformational changes.11
Featured super stable S protein trimer from ACROBiosystems
ACROBiosystems offers an extremely stable S protein trimer that can be used for vaccine and therapeutic development. In addition to the 6 prolines substitution, the trimer contains a T4 fibritin trimerization motif to stabilize the trimeric prefusion state. The furin cleavage site has also been mutated via two alanine substitutions.
ACROBiosystems has isolated high purity S trimer with high homogeneity. The S trimer particle structure was confirmed by utilizing negative stain electron microscopy and matches reports from the literature.
Furthermore, the performance testing of their Super stable S trimer with WT S trimer and 2 proline substituted S trimer along with S trimer from a competitor demonstrated that ACROBiosystems Super stable S trimer with 6 proline mutation has the highest signal to noise ratio.
This extremely high performance will improve vaccine immunogenicity evaluations and the development of diagnostic kits greatly.
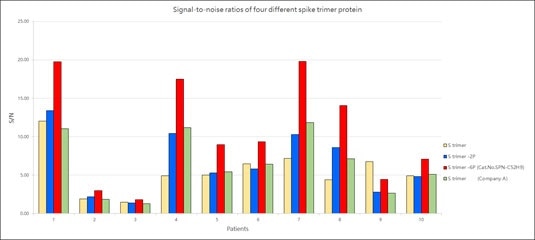
Figure 9. Signal-to-noise ratios of four different spike trimer protein. Image Credit: ACROBiosystems
References
- B, H.; H, G.; P, Z.; ZL, S., Characteristics of SARS-CoV-2 and COVID-19. Nature reviews. Microbiology 2020.
- E, P.; M, K.; U, G.; DH, H.; N, P.; F, C.; M, S.; S, A. K.; L, S., Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. The Lancet. Infectious diseases 2020, 20 (9).
- Z, Z.; X, L.; X, S.; W, W.; GA, M.; Y, Z., From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 2020, 21 (1).
- JM, W.; SE, D.; M, B.; K, S., Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Critical reviews in biochemistry and molecular biology 2008, 43 (3).
- T, T.; M, B.; JA, J.; GR, W.; S, D., Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research 2020, 178.
- Buchholz, U.; Bukreyev, A.; Yang, L.; Lamirande, E.; Murphy, B.; Subbarao, K.; Collins, P., Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proceedings of the National Academy of Sciences of the United States of America 2004, 101 (26).
- Traggia, i. E.; Becker, S.; Subbarao, K.; Kolesnikova, L.; Uematsu, Y.; Gismondo, M.; Murphy, B.; Rappuoli, R.; Lanzavecchia, A., An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature medicine 2004, 10 (8).
- J, S.; Y, W.; C, L.; G, Y.; Q, G.; A, A.; F, L., Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America 2020, 117 (21).
- DJ, B.; AG, W.; P, X.; C, R.; SR, M.; PB, R.; JJ, S.; SJ, G., Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588 (7837).
- R, H.; RJ, E.; K, M.; K, J.; V, S.; SMC, G.; M, K.; D, L.; R, P.; AL, H.; MJ, B.; BF, H.; P, A., Controlling the SARS-CoV-2 spike glycoprotein conformation. Nature structural & molecular biology 2020, 27 (10).
- CL, H.; JA, G.; JM, S.; AM, D.; HC, K.; K, J.; KC, L.; D, W.; AG, L.; Y, L.; CW, C.; PO, B.; CK, H.; NV, J.; J, L.-M.; AW, N.; J, P.; N, W.; D, A.; JJ, L.; GC, I.; JA, M.; IJ, F.; JS, M., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science (New York, N.Y.) 2020, 369 (6510).
- D, W.; N, W.; KS, C.; JA, G.; CL, H.; O, A.; BS, G.; JS, M., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020, 367 (6483).
- R, Y.; Y, Z.; Y, L.; L, X.; Y, G.; Q, Z., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 2020, 367 (6485).
- J, L.; J, G.; J, Y.; S, S.; H, Z.; S, F.; Q, Z.; X, S.; Q, W.; L, Z.; X, W., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581 (7807).
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L. D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Meer, F. v. d.; Kallio, K.; Kaya, T.; Anastasina, M.; Smura, T.; Levanov, L.; Szirovicza, L.; Tobi, A.; Kallio-Kokko, H.; Österlund, P.; Joensuu, M.; Meunier, F. A.; Butcher, S. J.; Winkler, M. S.; Mollenhauer, B.; Helenius, A.; Gokce, O.; Teesalu, T.; Hepojoki, J.; Vapalahti, O.; Stadelmann, C.; Balistreri, G.; Simons, M., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370 (6518), 856-860.
- C, W.; L, W.; Q, Y.; X, W.; J, Z.; X, Y.; Y, Z.; C, F.; D, L.; Y, D.; J, S.; J, G.; X, Y.; Y, W.; X, W.; J, L.; H, Y.; H, L.; Z, Z.; R, W.; P, D.; Y, Z.; F, Y.; W, Z.; N, W.; Y, P.; H, L.; J, F.; C, Q.; W, C.; Q, G.; R, Z.; Y, C.; H, Z., HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nature metabolism 2020, 2 (12).
- Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; Zhang, J.; Shen, G.; Jiang, X.; Yang, J.; Zheng, X.; Xu, J.; Zhang, C. C.; Lan, F.; Qu, D.; Zhao, Y.; Xu, G.; Xie, Y.; Luo, M.; Lu, Z., Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv 2020.
- Wang, S., Qiu, Z., Hou, Y. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31, 126–140 (2021).
- L, Z.; CB, J.; H, M.; A, O.; H, P.; BD, Q.; ES, R.; A, P.; A, V.; MS, S.; W, L.; T, I.; C, R.; M, F.; H, C., SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nature communications 2020, 11 (1).
- L, Y.; X, W.; KE, P.; C, T.-T.; TP, N.; Y, W.; A, B.; WE, D.; A, D.; C, C.; K, V.; SB, E.; SF, S.; JE, L.; JB, M.; A, R.; A, B.; PC, S.; CA, K.; NV, D.; K, S.; J, L., Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183 (3).
- Zhang, W.; Davis, B. D.; Chen, S. S.; Martinez, J. M. S.; Plummer, J. T.; Vail, E., Emergence of a novel SARS-CoV-2 strain in Southern California, USA. Preprint 2021.
- Wang, P.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P. D.; Graham, B. S.; Mascola, J. R.; Chang, J. Y.; Yin, M. T.; Sobieszczyk, M.; Kyratsous, C. A.; Shapiro, L.; Sheng, Z.; Nair, M. S.; Huang, Y.; Ho, D. D., Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv 2021.
- PANGO lineages. https://cov-lineages.org/global_report.html.
- Wu, K.; Werner, A. P.; Moliva, J. I.; Koch, M.; Choi, A.; Stewart-Jones, G. B. E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B. S.; Carfi, A.; Corbett, K. S.; Seder, R. A.; Edwards, D. K., mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021.
- Xie, X.; Zou, J.; Fontes-Garfias, C. R.; Xia, H.; Swanson, K. A.; Cutler, M.; Cooper, D.; Menachery, V. D.; Weaver, S.; Dormitzer, P. R.; Shi, P.-Y., Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021.
- Novavax, Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. 2021.
- Greaney, A. J.; Loes, A. N.; Crawford, K. H. D.; Starr, T. N.; Malone, K. D.; Chu, H. Y.; Bloom, J. D., Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. 2021.
- Luan, B.; Wang, H.; Huynh, T., Molecular Mechanism of the N501Y Mutation for Enhanced Binding between SARS-CoV-2’s Spike Protein and Human ACE2 Receptor. bioRxiv 2021.
- RL, G.; A, N.; P, C.; J, B.; B, H.; J, M.; G, H.; J, C.; B, M.; V, S.; I, S.; P, K.; AC, A.; J, V. N.; KL, C.; M, D.; G, O.; AE, S.; TR, H.; PJ, E.; RE, H.; NL, K.; J, S.; DR, P.; P, K.; L, S.; DM, S., Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021.
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.