Over 1.8 million infections and over 118 thousand deaths have resulted from the emergence of SARS-CoV-2 and this number is still growing. SARS-CoV-2 is similar to SARS-CoV and MERS-CoV. It is highly transmissible from infected individuals to healthy ones, even without symptoms, which can cause extremely severe respiratory symptoms.
The requirement to develop vaccines quickly. In addition to therapeutics against SARS-CoV-2 at a time of the explosion is crucial. Coronavirus entry into the host cell is mediated through the transmembrane spike (S) glycoprotein which forms homotrimers protruding from the viral surface.
In addition to the focus of therapeutic and vaccine design, this S glycoprotein is the key target of neutralizing antibodies upon infection. S trimers are extensively decorated with N-linked glycans which are vital for proper folding and for modulating accessibility to host proteases.
In this article, a few recent studies are outlined in order to look at the more scientific details about the glycosylation of the S protein and the importance of it.
1. Site-specific analysis of the SARS-CoV-2 glycan shield[1]
Crispin team expressed and purified recombinant SARS-CoV-2 S protein successfully from HEK293F cells. Using a mass spectrometer, they examined the composition of site-specific N-linked glycerin addition to the degree of sequon occupancy on the protein.
Each of the 22 glycans on the SARS-CoV-2 S protein were identified, and 18 of those N-glycosides were conserved between SARS-CoV and SARS-CoV-2 S proteins, as seen in Figure 1.
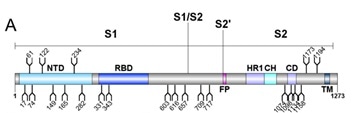
Figure 1. Schematic representation of SARS-CoV-2 S glycoprotein. Image Credit: ACROBiosystems
The abundances of each glycan are summed into oligomannose-, hybrid, and complex-type glycosylation, showing the extensive heterogeneity in N-glycan composition and site occupancy.
Furthermore, as seen in Figure 2, the diverse signals which come from heterogeneous complex-type glycosylation are simplified by the summation of glycan intensities into a more limited variety of structural categories.
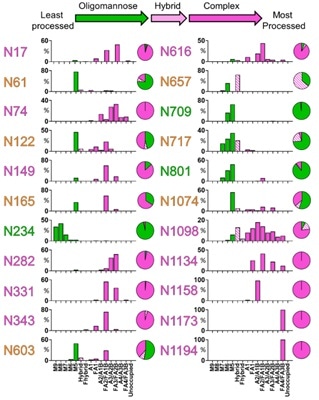
Figure 2. Site-specific N-linked glycosylation of SARS-CoV-2 S glycoprotein. Image Credit: ACROBiosystems
2. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins using high-resolution mass spectrometry[2]
Recently, a team from West China Hospital of Sichuan University published their findings. They characterized the site-specific N-glycosylation of HEK293 cell expressed SARS-CoV-2 S1 protein and insect cell expressed S protein.
They carried out stepped collision energy (SCE) mass spectrometry following digestion with two complementary proteases to cover all potential N-glycosylation sequons and integrated N-glycoproteomics analysis.
This showed the N-glycosylation profile of SARS-CoV-2 S proteins at the levels of intact N-glycopeptides and glycosites, in addition to the glycan composition and site-specific number of glycans, as seen in Figure 3.
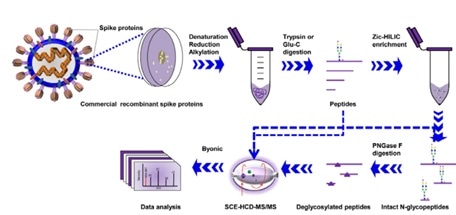
Figure 3. Workflow for site-specific N-glycosylation characterization of recombinant SARS-CoV-2 S proteins using two complementary proteases for digestion and simultaneous N-glycoproteomics analysis. Image Credit: ACROBiosystems
Each of the 22 potential canonical N-glycosites were identified in the S protein protomer. Notably, the human cell-produced protein possessed up to 140 N-glycans, most of which are complex type, whereas insect cell-produced SARS-CoV-2 S protein had 38 N-glycans, which were mostly high-mannose type N-glycans, as seen in Figure 4.
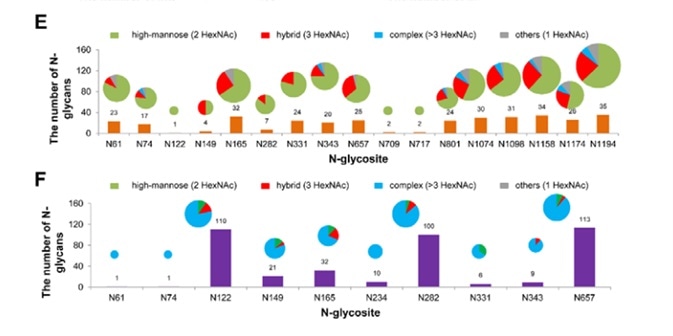
Figure 4. Site-specific N-glycosylation of recombinant SARS-CoV-2 S proteins. Image Credit: ACROBiosystems
This N-glycosylation profiling and determination of differences between distinct expression systems could shed light on the infection mechanism and promote the development of targeted therapeutics and vaccines.
3. Deducing the N- and O- glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2[3]
Azadi’s team from the University of Georgia published their findings on bioRxiv. They digested the recombinant SARS-CoV-2 subunit S1 and S2 expressed in HEK293 cells into the glycopeptides. Next, to analyze these glycopeptides, high resolution LC-MS/MS was utilized.
Using this method, they were able to identify partial N-glycan occupancy on 17 out of 22 N-glycosylation sites with a combination of high mannose and complex type, but no hybrid-type glycans, as seen in Figure 5.
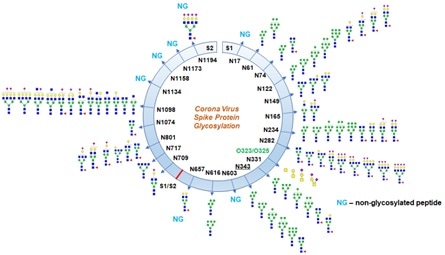
Figure 5. Glycosylation profile on coronavirus SARS-CoV-2 characterized by high-resolution LCMS/MS. Image Credit: ACROBiosystems
It is worth noting that they identified two unexpected O-glycosylation modifications on the receptor binding domain (RBD) of spike protein subunit S1. Even though O-glycosylation has been expected on the spike protein of SARS-Cov-2, this is the first report of the site of O-glycosylation and identity of the O-glycans attached on the subunit S1.
4. Structural and functional analysis of a potent sarbecovirus neutralizing antibody[4]
Vir Biotechnology worked with the University of Washington to examine the potential cross reactivity between the neutralizing monoclonal antibodies identified from an individual infected with SARS-CoV in 2003 and SARS-CoV-2.
As seen in Figure 6, by engaging the S receptor binding domain, mAb S309 potently neutralized SARS-CoV-2, SARS-CoV pseudoviruses, in addition to the authentic SARS-CoV-2.
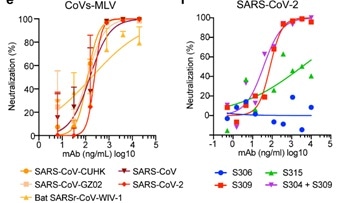
Figure 6. Neutralization of pseudoviruses (bearing S protein) and authentic SARS-CoV-2 by an isolated antibody named S309 from a SARS survivor. Image Credit: ACROBiosystems
It was demonstrated via cryo-electron microscopic analysis and different binding assays that S309 recognized a glycan-containing epitope, which is conserved within the sarbecovirus subgenus, without competing with receptor binding. This is shown in Figure 7.
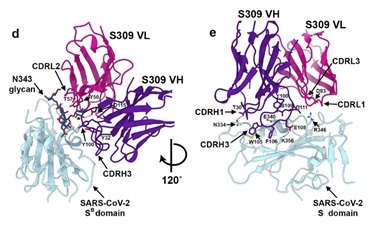
Figure 7. Closeup view of the S309 epitope showing the contacts formed with the core fucose (labeled with a star) and the core N-acetyl-glucosamine of the oligosaccharide at position N343. Image Credit: ACROBiosystems
The glycan recognition of S309 shows the importance of the naturally decorated N-glycans in SARS-CoV-2 S proteins.
5. 3D models of glycosylated SARS-CoV-2 spike protein suggest challenges and opportunities for vaccine development[5]
A study regarding the 3D models of glycosylated SARS-CoV-2 spike protein was reported by a group of researchers from the University of Georgia. Their molecular dynamics simulation result showed that glycans shield around 40 % of the underlying protein surface of the S glycoprotein from antibody recognition, but the efficacy of antisera should not be affected.
The largest and most accessible antigenic surface in the S protein is made up of the ACE2 binding domain. This suggests that, as long as the virus continues to target the same host receptor, a vaccine with this epitope could be effective, as seen in Figure 8.
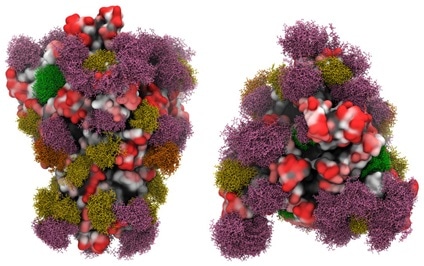
Figure 8. Side view (left panels) and Top view (right panels) of the SARS-CoV-2 S glycoprotein with site-specific glycosylation. Image Credit: ACROBiosystems
Additionally, efficacy could benefit from ensuring that the production technique leads to glycosylation profiles which match those of the circulating virus, or by engineering a vaccine to avoid glycosylated sequences.
Glycosylation of the S protein is also complex and important, as we can see from the previous studies. Different expressing systems can generate proteins with completely different glycosylation results in terms of the type and composition of glycan.
Crucially, the glycosylation can influence the vaccine and therapeutics development, which should draw much more attention from scientists around the globe. The drug industry is hoping to shorten the time it takes to get a vaccine and the therapeutics to market which is typically between 10 and 15 years, to within the next year. Many challenges can be avoided by selecting the right protein reagent.
Based on their HEK293 protein expression platform, ACROBiosystems has developed a number of SARS-CoV-2 proteins. The state of their mammalian cell expressed recombinant proteins is nearer to that of the protein in humans including the glycosylation level.
Their data showed that SARS-CoV-2 S proteins expressed by different expressing systems can lead to the different binding affinities to ACE2. This could be due to the different post translational modifications, particularly the glycosylation between two different expressing systems.
Other than the S protein, ACROBiosystems developed S, S1, S-RBD and ACE2 proteins, which are verified by Biacore and ELISA assays.
Intended for the development of the vaccine, therapeutics and diagnostic cassettes, in terms of mimicking the condition in blood circulation these HEK293 expressed SARS-CoV-2 proteins do have benefits.
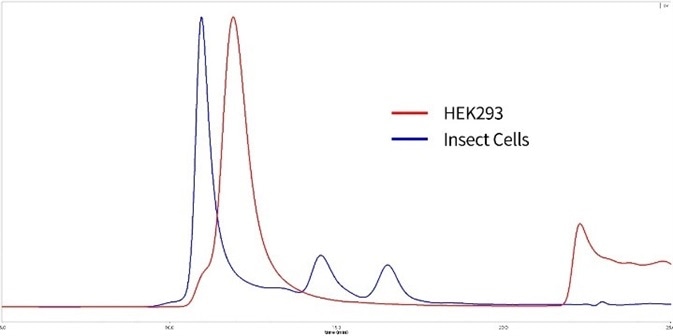
Figure 9. SEC-MALS of HEK293 expressed and insect cell expressed S proteins. Red: HEK293 expressed S protein is verified to be trimer with 85% purity. MW is 600 to 660 kDa. Blue: Insect cell expressed S protein is in high polymerization state. Image Credit: ACROBiosystems
(Related ACRO product: SARS-CoV-2 (COVID-19) S protein (R683A, R685A), His Tag, active trimer, HEK 293)
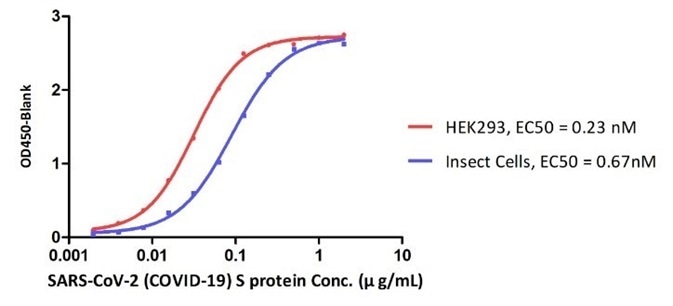
Figure 10. ELISA Binding affinity between ACE2 protein and S proteins expressed by HEK293 cell and insect cell respectively. Image Credit: ACROBiosystems
References
- Yasunori Watanabe et al. Site-specific analysis of the SARS-CoV-2 glycan shield. bioRxiv preprint doi: https://doi.org/10.1101/2020.03.26.010322
- Yong Zhang et al. Site-specific N-glycosylation Characterization of Recombinant SARS-CoV-2 Spike Proteins using High-Resolution Mass Spectrometry. bioRxiv preprint doi: https://doi.org/10.1101/2020.03.28.013276.
- Asif Shajahan et al. Deducing the N- and O- glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. bioRxiv preprint
- Dora Pinto et al. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv preprint doi: https://doi.org/10.1101/2020.04.07.023903.
- Oliver C. Grant et al. 3D Models of glycosylated SARS-CoV-2 spike protein suggest challenges and opportunities for vaccine development. bioRxiv preprint doi: http://biorxiv.org/cgi/content/short/2020.04.07.030445.
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
- Walls et al., Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell (2020).
- Walls, A.C. et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 23, 899–905. (2016).
- Walls, A.C. et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 114, 11157–11162. (2017).
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.