As reports suggest, reduced vaccine and antibody therapeutic efficacy and higher transmissibility and in these strains, mutations in the RBD have become a cause of concern.
Random mutations of the viral genome are inevitable as SARS-CoV-2 spreads around the world. Mutations in and outside the RBD domain can alter the conformation of S protein significantly.
This must be continuously monitored for effect in transmissibility and severity of COVID-19. In the course of this pandemic, GISAID is tracking new mutation/clades of SARS-CoV-2.
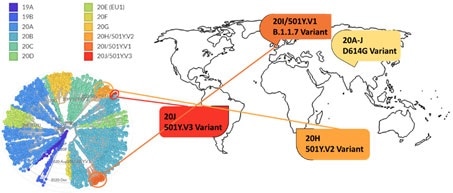
Figure 14. Phylogenetic clades of SARS-CoV-2 mutations with key variants around the world highlighted. Image Credit: ACROBiosystems
Next strain visualization of the SARS-CoV-2 mutation exhibits numerous clades of mutants emerging in 2019 and 2020, seen in Figure 14. Yet, mutations which result in a gain of functions are particularly dangerous as they could increase disease severity and viral transmissibility.
The D614G variant is the dominant mutation found in almost all SARS-CoV-2 infections and has been attributed to the growth in SARS-CoV-2 transmission.
Numerous explanations for high transmissibility of the D614G variant have been demonstrated and include improvement in the assembly of functional S protein on virion19 and equilibrium shift of RBD from 47% open to 95% open conformation in the mutant20.
Three new variants have recently emerged: 501Y.V1/B.1.1.7 from the UK, 501Y.V2/P.1 from Brazil, and 501Y.V2/B.1.351 variant from South Africa. A US variant originating on the west coast (B.1.429) has been more recently identified and suspected in a quick rise of cases.21
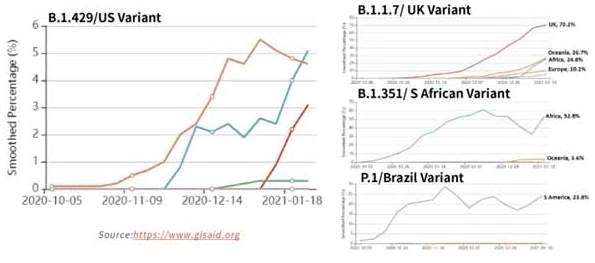
Figure 15. Tracking of variant percentages in SARS-CoV-2 samples as of Jan 28. Image Credit: ACROBiosystems
Due to the potential resistance to vaccine-induced/therapeutic neutralizing antibodies and suspected rise in transmissibility, the spread of these strains globally, seen in Figure 15, has been alarming. Table 3 shows the mutation details for the variants.
Table 3. Major SARS-CoV-2 variants in circulation worldwide21-23. Bold mutations are in the RBD and red-colored mutations lie within the RBM. Source: ACROBiosystems
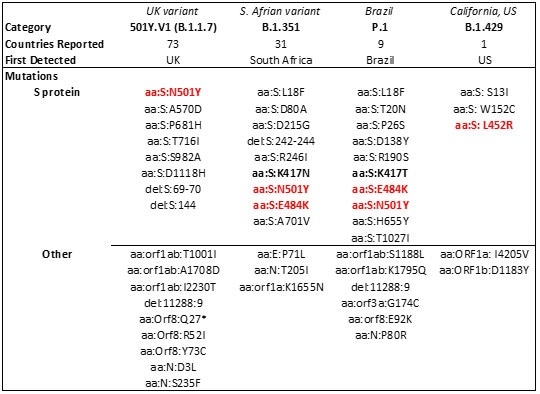
As reports indicate reduced vaccine and antibody therapeutic efficacy and higher transmissibility in these strains, mutations in the RBD have become a cause of concern.
Recent reports indicate that the B.1.1.7 variant is resistant to neutralization by antibodies targeting NTD of S protein and partially resistant to RBD mutants while showing modest resistance in convalescent plasma and vaccinated sera.22
The B.1.351 has exhibited higher resistance to both NTD and RBD antibodies and significant resistance (11-33 fold) in convalescent plasma and vaccine sera (6.5-8.6).
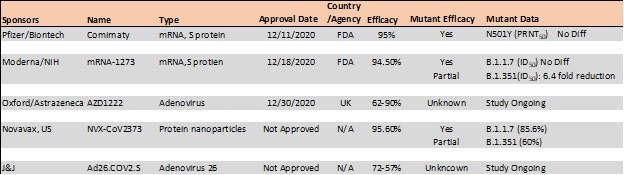
In results with Moderna, Pfizer/BioNTech, and Novavax vaccines, the antibody titer exhibits no difference against B.1.1.7 variant while lowered efficacy has been seen against B.1.351 variant 24, 25.
Table 4. Vaccine efficacy analysis against emerging strains24-26. Source: ACROBiosystems
The E484K mutation, which is present in both S. Africa and Brazil variants, is thought to contribute to resistance where neutralization decrease >10 fold was seen in some convalescent serum antibodies.27
Another mutant, N501Y, found in the UK, S African, and Brazil variants is thought to enhance the transmissibility of SARS-CoV-2 via strengthening ACE2 binding.28 Further to effect in vaccination, these emerging variants have been discovered to lower the potency of current antibody therapies against COVID-19.
Two monoclonal antibody cocktails have been tested against these variants and have received emergency use authorization from the FDA. They are from Eli Lilly (bamlanivimab/estesevimab) and Regeneron (Casirivimab/imdevimab).
For Eli Lilly’s antibodies, bamlanivimab was susceptible to the E484K mutation in RBD, which resulted in an over 100-fold reduction in neutralizing activity while estesevimab retained activity with only a 4.4-fold decrease in neutralizing ability.29
Regeneron’s casirivimab and bamlanivimab both exhibited little to no change in potency against the B.1.1.7 variant whilst exhibiting complete or extremely abolished activity against B.1.347 variants.22
For these variants, ACROBiosystems is working to develop a collection of recombinant antigens. The reagents can be utilized to assess the efficacy of the vaccination and antibodies.
Newly launched product: Neutralizing antibody titer kit accelerating vaccines and diagnostics development
A number of serological tests have been developed by ACROBiosystems to track and fight the surging COVID-19 spread. To establish the efficacy of vaccines and for a diagnostic measure of past infections, testing the levels of neutralizing antibodies against S protein in serum samples is vital.
With the rise of mutant variants and the start of large-scale immunization with multiple vaccines, close monitoring of neutralizing antibodies in the population and the effect of virus mutation is needed for effective immunization campaigns. ACROBiosystems provides neutralization kits and IgG, IgM titer detection kits.
The neutralization kit from ACROBiosystems demonstrated consistent detection of neutralizing antibodies against numerous dilutions of COVID-19 convalescent serum.
ACROBiosystems’ serology kits provide a better understanding of neutralizing antibody mechanisms and help researchers develop strategies to improve vaccines and diagnostics, particularly against emerging SARS-CoV-2 mutant strains.
References
- B, H. H, G.; P, Z.; ZL, S., Characteristics of SARS-CoV-2 and COVID-19. Nature reviews. Microbiology 2020.
- E, P. M, K.; U, G. DH, H. N, P.; F, C. M, S. S, A. K.; L, S., Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. The Lancet. Infectious diseases 2020, 20 (9).
- Z, Z. X, L.; X, S. W, W. GA, M.; Y, Z., From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 2020, 21 (1).
- JM, W. SE, D.; M, B.; K, S., Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Critical reviews in biochemistry and molecular biology 2008, 43 (3).
- T, T. M, B.; JA, J. GR, W.; S, D., Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research 2020, 178.
- Buchholz, U. Bukreyev, A.; Yang, L. Lamirande, E. Murphy, B.; Subbarao, K.; Collins, P., Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proceedings of the National Academy of Sciences of the United States of America 2004, 101 (26).
- Traggia, i. E. Becker, S.; Subbarao, K. Kolesnikova, L. Uematsu, Y. Gismondo, M.; Murphy, B. Rappuoli, R. Lanzavecchia, A., An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature medicine 2004, 10 (8).
- J, S. Y, W.; C, L. G, Y. Q, G.; A, A.; F, L., Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America 2020, 117 (21).
- DJ, B. AG, W.; P, X. C, R. SR, M.; PB, R. JJ, S.; SJ, G., Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588 (7837).
- R, H. RJ, E. K, M.; K, J. V, S. SMC, G.; M, K. D, L. R, P.; AL, H. MJ, B. BF, H.; P, A., Controlling the SARS-CoV-2 spike glycoprotein conformation. Nature structural & molecular biology 2020, 27 (10).
- CL, H. JA, G. JM, S.; AM, D. HC, K. K, J.; KC, L. D, W. AG, L.; Y, L. CW, C. PO, B.; CK, H. NV, J. J, L.-M.; AW, N. J, P. N, W.; D, A. JJ, L. GC, I.; JA, M. IJ, F.; JS, M., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science (New York, N.Y.) 2020, 369 (6510).
- D, W. N, W. KS, C. JA, G.; CL, H. O, A. BS, G.; JS, M., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020, 367 (6483).
- R, Y. Y, Z. Y, L.; L, X. Y, G.; Q, Z., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 2020, 367 (6485).
- J, L. J, G. J, Y.; S, S. H, Z. S, F.; Q, Z. X, S. Q, W.; L, Z.; X, W., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581 (7807).
- Cantuti-Castelvetri, L.; Ojha, R. Pedro, L. D. Djannatian, M. Franz, J.; Kuivanen, S. Meer, F. v. d. Kallio, K. Kaya, T.; Anastasina, M. Smura, T. Levanov, L.; Szirovicza, L. Tobi, A. Kallio-Kokko, H. Österlund, P. Joensuu, M.; Meunier, F. A. Butcher, S. J. Winkler, M. S. Mollenhauer, B. Helenius, A.; Gokce, O. Teesalu, T. Hepojoki, J.; Vapalahti, O. Stadelmann, C. Balistreri, G.; Simons, M., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370 (6518), 856-860.
- C, W. L, W. Q, Y.; X, W. J, Z. X, Y.; Y, Z. C, F. D, L.; Y, D. J, S. J, G.; X, Y. Y, W. X, W.; J, L. H, Y. H, L.; Z, Z. R, W. P, D.; Y, Z. F, Y. W, Z.; N, W. Y, P. H, L.; J, F. C, Q. W, C.; Q, G. R, Z. Y, C.; H, Z., HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nature metabolism 2020, 2 (12).
- Gu, Y. Cao, J. Zhang, X.; Gao, H. Wang, Y. Wang, J.; Zhang, J. Shen, G. Jiang, X.; Yang, J. Zheng, X. Xu, J.; Zhang, C. C. Lan, F. Qu, D.; Zhao, Y. Xu, G. Xie, Y.; Luo, M.; Lu, Z., Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv 2020.
- Wang, S., Qiu, Z., Hou, Y. et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31, 126–140 (2021).
- L, Z. CB, J. H, M.; A, O. H, P. BD, Q.; ES, R. A, P. A, V.; MS, S. W, L. T, I.; C, R. M, F.; H, C., SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nature communications 2020, 11 (1).
- L, Y. X, W. KE, P.; C, T.-T. TP, N. Y, W.; A, B. WE, D. A, D.; C, C. K, V. SB, E.; SF, S. JE, L. JB, M.; A, R. A, B. PC, S.; CA, K. NV, D. K, S.; J, L., Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183 (3).
- Zhang, W. Davis, B. D. Chen, S. S.; Martinez, J. M. S. Plummer, J. T.; Vail, E., Emergence of a novel SARS-CoV-2 strain in Southern California, USA. Preprint 2021.
- Wang, P. Liu, L. Iketani, S.; Luo, Y. Guo, Y. Wang, M.; Yu, J. Zhang, B. Kwong, P. D.; Graham, B. S. Mascola, J. R. Chang, J. Y. Yin, M. T.; Sobieszczyk, M. Kyratsous, C. A. Shapiro, L.; Sheng, Z. Nair, M. S. Huang, Y.; Ho, D. D., Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv 2021.
- PANGO lineages. https://cov-lineages.org/global_report.html.
- Wu, K. Werner, A. P. Moliva, J. I. Koch, M.; Choi, A. Stewart-Jones, G. B. E. Bennett, H. Boyoglu-Barnum, S. Shi, W.; Graham, B. S. Carfi, A. Corbett, K. S. Seder, R. A.; Edwards, D. K., mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021.
- Xie, X. Zou, J. Fontes-Garfias, C. R. Xia, H.; Swanson, K. A. Cutler, M. Cooper, D.; Menachery, V. D. Weaver, S. Dormitzer, P. R.; Shi, P.-Y., Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021.
- Novavax, Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. 2021.
- Greaney, A. J. Loes, A. N. Crawford, K. H. D. Starr, T. N.; Malone, K. D. Chu, H. Y.; Bloom, J. D., Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. 2021.
- Luan, B. Wang, H.; Huynh, T., Molecular Mechanism of the N501Y Mutation for Enhanced Binding between SARS-CoV-2’s Spike Protein and Human ACE2 Receptor. bioRxiv 2021.
- RL, G. A, N.; P, C. J, B. B, H.; J, M. G, H. J, C.; B, M. V, S. I, S.; P, K. AC, A. J, V. N.; KL, C. M, D. G, O.; AE, S. TR, H. PJ, E.; RE, H. NL, K. J, S.; DR, P. P, K. L, S.; DM, S., Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021.
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.